4BIQ
 
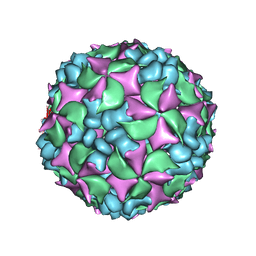 | | Homology model of coxsackievirus A7 (CAV7) empty capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
6RUS
 
 | | Structure of a functional properdin monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, SULFATE ION, ... | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2019-05-29 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
6TLZ
 
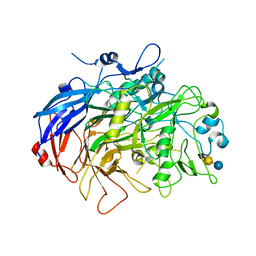 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 3'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
6T5B
 
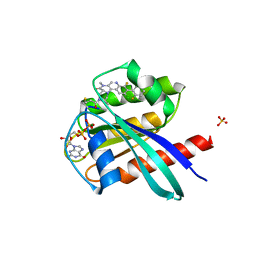 | | KRasG12C ligand complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T5U
 
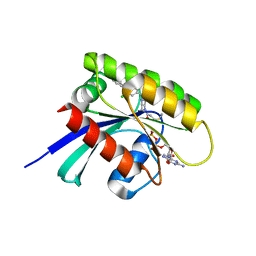 | | KRasG12C ligand complex | | Descriptor: | 1-[(7R)-16-chloro-15-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,12-triazatetracyclo[8.8.0.02,7.013,18]octadeca-1(10),11,13,15,17-pentaen-5-yl]prop-2-en-1-one, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T6Q
 
 | | Crystal structure of Toxoplasma gondii Morn1 (extended conformation). | | Descriptor: | Membrane occupation and recognition nexus protein MORN1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T5L
 
 | |
3P0G
 
 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
6T5V
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
4CIK
 
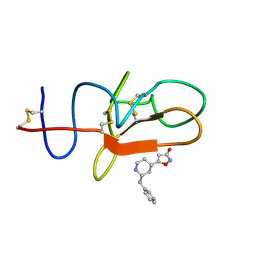 | | plasminogen kringle 1 in complex with inhibitor | | Descriptor: | 5-[(2R,4S)-2-(phenylmethyl)piperidin-4-yl]-1,2-oxazol-3-one, PLASMINOGEN | | Authors: | Xue, Y, Johansson, C, Cheng, L, Pettersen, D, Gustafsson, D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of the Fibrinolysis Inhibitor Azd6564, Acting Via Interference of a Protein-Protein Interaction.
Acs Med.Chem.Lett., 5, 2014
|
|
4CL6
 
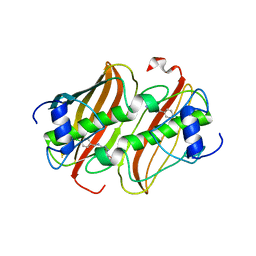 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N-(4- Chlorobenzyl)-3-(2-furyl)-1H-1,2,4-triazol-5-amine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-(4-chlorobenzyl)-5-(furan-2-yl)-1H-1,2,4-triazol-3-amine | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Substrate Mimic Allows High Throughput Assay of the Faba Protein and Consequently the Identification of a Novel Inhibitor of Pseudomonas Aeruginosa Faba.
J.Mol.Biol., 428, 2016
|
|
4CQH
 
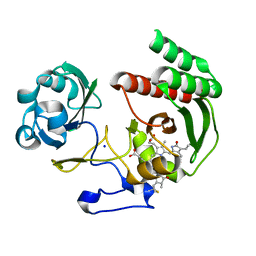 | | Structure of Infrared Fluorescent Protein IFP2.0 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME, SODIUM ION | | Authors: | Lafaye, C, Yu, D, Noirclerc-Savoye, M, Shu, X, Royant, A. | | Deposit date: | 2014-02-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An Improved Monomeric Infrared Fluorescent Protein for Neuronal and Tumour Brain Imaging.
Nat.Commun., 5, 2014
|
|
6RR3
 
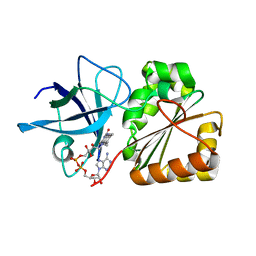 | |
6RIU
 
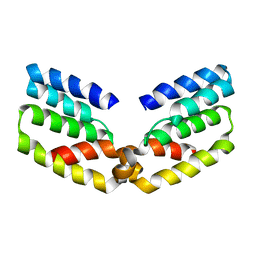 | |
6RJ1
 
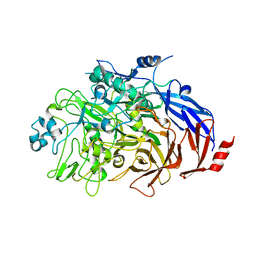 | |
4CTN
 
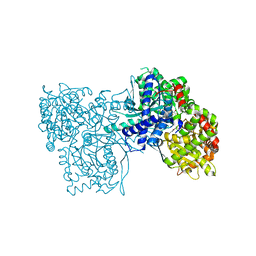 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DIMETHYL SULFOXIDE, GLYCOGEN PHOSPHORYLASE, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
4D45
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-bromo- 2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
6RVB
 
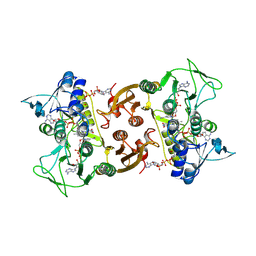 | | NADH-dependent Coenzyme A Disulfide Reductase soaked with NADH | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase, ... | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6RFO
 
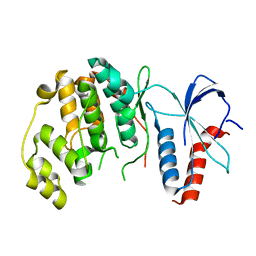 | | ERK2 MAP kinase with the activation loop of p38alpha | | Descriptor: | Mitogen-activated protein kinase 1,Mitogen-activated protein kinase 14,Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Eitan-Wexler, M. | | Deposit date: | 2019-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop.
J.Biol.Chem., 295, 2020
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
4A7I
 
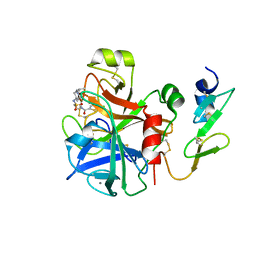 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ALZ
 
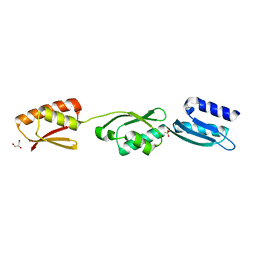 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
6TNC
 
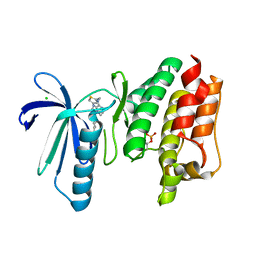 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 46 | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
6T68
 
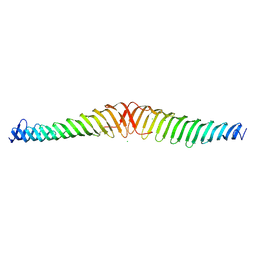 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
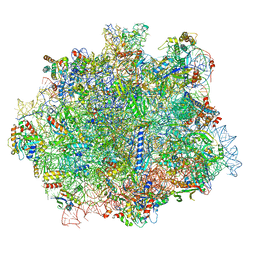
6RI5
 
 | |
