6Z9Z
 
 | | Atomic resolution X-ray structure of the Uridine phosphorylase from Vibrio cholerae on crystals grown under microgravity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Eistrikh-Heller, P.A, Rubinsky, S.V, Samygina, V.R, Gabdulkhakov, A.G, Lashkov, A.A. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallization in Microgravity and the Atomic-Resolution Structure of Uridine Phosphorylase from Vibrio cholerae
Crystallography Reports, 66, 2021
|
|
3G71
 
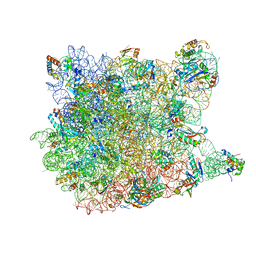 | | Co-crystal structure of Bruceantin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
5M99
 
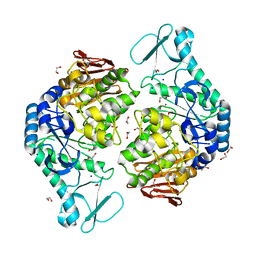 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
3I56
 
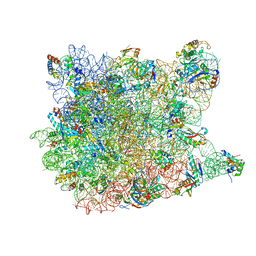 | | Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
6GZ8
 
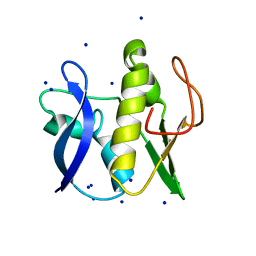 | | First GerMN domain of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
4N3P
 
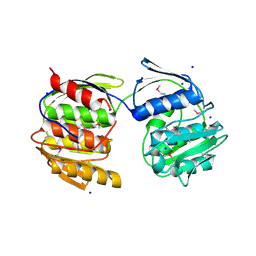 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
6XNE
 
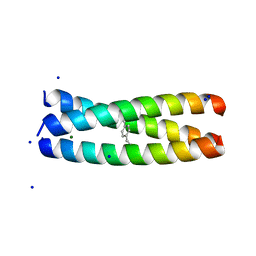 | |
7D3U
 
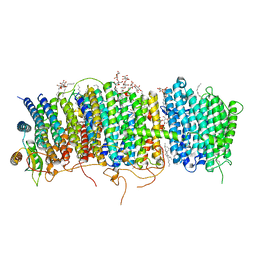 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y8O
 
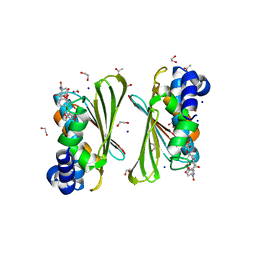 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
7XVV
 
 | |
7XVW
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVU
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7BOO
 
 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
7BOP
 
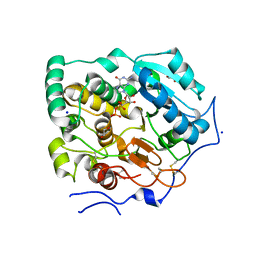 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
5AGV
 
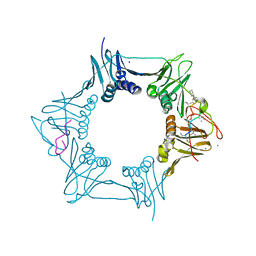 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, CYCLOHEXYL GRISELIMYCIN, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
4XTN
 
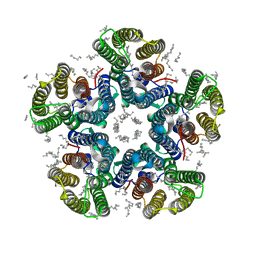 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 4.9 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6MLT
 
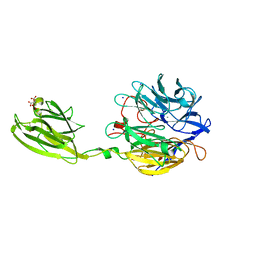 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | Descriptor: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
5BS4
 
 | | HIV-1 wild Type protease with GRL-047-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, 4-amino sulfonamide derivative) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
5B1F
 
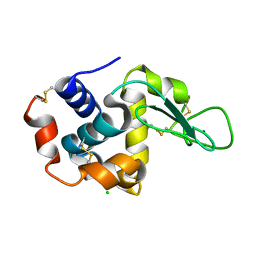 | |
5B43
 
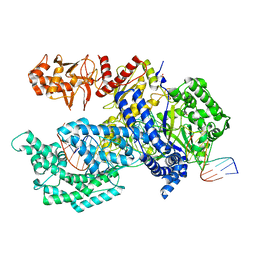 | | Crystal structure of Acidaminococcus sp. Cpf1 in complex with crRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cpf1, DNA (34-MER), ... | | Authors: | Yamano, T, Nishimasu, H, Hirano, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA
Cell, 165, 2016
|
|
5B6L
 
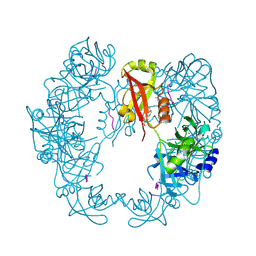 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
4XN2
 
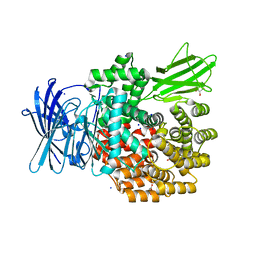 | |
5BOL
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dGTP analog | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BPC
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dATP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BPM
 
 | |
