2AGV
 
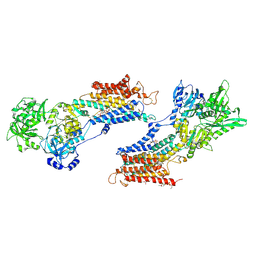 | | Crystal structure of the SR CA2+-ATPASE with BHQ and TG | | Descriptor: | 2,5-DITERT-BUTYLBENZENE-1,4-DIOL, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Obara, K, Norimatsu, Y. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inaugural Article: Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5QQO
 
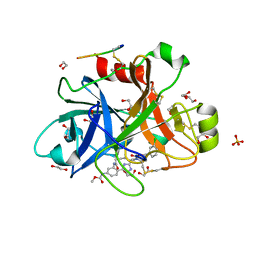 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(5E,8S)-8-[(6R)-6-(3-chlorophenyl)-2-oxo-1,3-oxazinan-3-yl]-2-oxo-1,3,4,7,8,10-hexahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, MET-ASP-ASP-ASP-ASP-LYS-MET-ASP-ASN-GLU-CYS-THR-THR-LYS-ILE-LYS-PRO-ARG, ... | | Authors: | Sheriff, S. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based design of macrocyclic factor XIa inhibitors: Discovery of cyclic P1 linker moieties with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3TIW
 
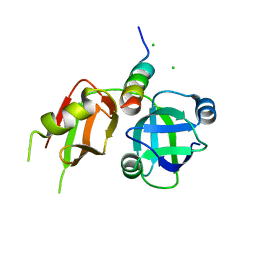 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
6GJW
 
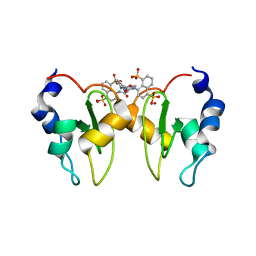 | | Structure of XIAP-BIR1 domain in complex with an NF023 analog | | Descriptor: | 4-[[3-[[3-[(4,8-disulfonatonaphthalen-1-yl)carbamoyl]phenyl]carbamoylamino]phenyl]carbonylamino]naphthalene-1,5-disulfonate, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Sorrentino, L, Cossu, F, Malkoc, B, Zaffaroni, M, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
6QI7
 
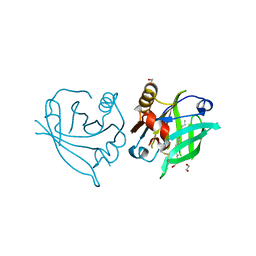 | | Engineered beta-lactoglobulin: variant L39Y in complex with endogenous ligand | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, PALMITIC ACID | | Authors: | Loch, J.I, Siuda, M.K, Lewinski, K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design approach to rational site-directed mutagenesis of beta-lactoglobulin.
J.Struct.Biol., 210, 2020
|
|
1R7I
 
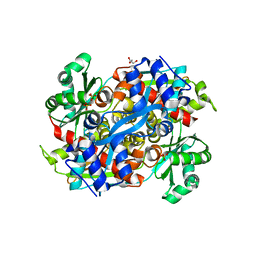 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
4DQD
 
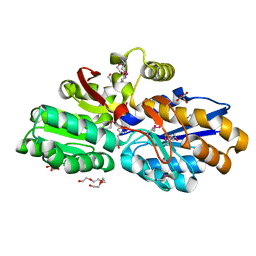 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4OJ4
 
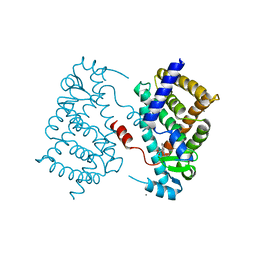 | |
4FB0
 
 | |
6WW4
 
 | | Crystal structure of HERC2 ZZ domain in complex with histone H3 tail | | Descriptor: | GLYCEROL, Histone H3.1,E3 ubiquitin-protein ligase HERC2, ZINC ION | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
1MOD
 
 | |
1MOB
 
 | |
3GM2
 
 | |
2O27
 
 | |
4FAX
 
 | |
4EYQ
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4JSA
 
 | | Benzenesulfonamide complexed with hCAII H94D | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, SULFATE ION, ... | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
2F7O
 
 | | Golgi alpha-mannosidase II complex with mannostatin A | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis of the Inhibition of Golgi alpha-Mannosidase II by Mannostatin A and the Role of the Thiomethyl Moiety in Ligand-Protein Interactions.
J.Am.Chem.Soc., 128, 2006
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
6AAO
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAP
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZaeSeCys | | Descriptor: | 3-[(2-{[(benzyloxy)carbonyl]amino}ethyl)selanyl]-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAD
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mTmdZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-[({3-[3-(trifluoromethyl)-3H-diaziren-3-yl]phenyl}methoxy)carbonyl]-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAQ
 
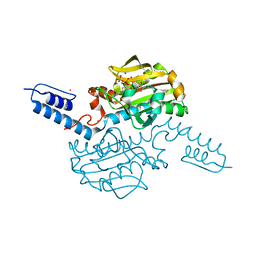 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with BCNLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-({[(1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl]methoxy}carbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAN
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mEtZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
1MOC
 
 | |
