2L6B
 
 | |
7SL3
 
 | |
7SL4
 
 | |
7SL1
 
 | |
7S5O
 
 | |
5HAL
 
 | |
6DVL
 
 | |
6DVN
 
 | |
6DJC
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5H6H
 
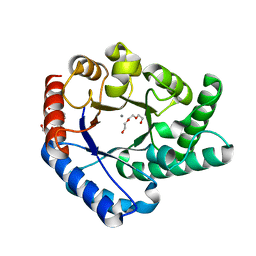 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ribulose 3-Epimerase with Mn2+ | | Descriptor: | MANGANESE (II) ION, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Cao, T.-P, Choi, J.M, Shin, S.M, Le, D.W, Lee, S.H. | | Deposit date: | 2016-11-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
2LT1
 
 | |
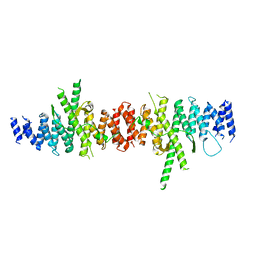
5HAS
 
 | |
4BPH
 
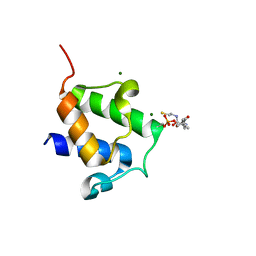 | | High resolution crystal structure of Bacillus subtilis DltC | | Descriptor: | 4'-PHOSPHOPANTETHEINE, D-ALANINE--POLY(PHOSPHORIBITOL) LIGASE SUBUNIT 2, MAGNESIUM ION | | Authors: | Zimmermann, S, Neumann, P, Stubbs, M.T. | | Deposit date: | 2013-05-26 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of the D-Alanyl Carrier Protein (Dcp) Dltc from Bacillus Subtilis Reveal Equivalent Conformations of Apo- and Holo-Forms
FEBS Lett., 589, 2015
|
|
4C61
 
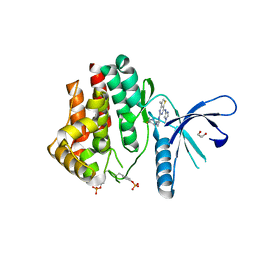 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | ACETATE ION, N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-7-methyl-N4-(1-methylimidazol-4-yl)thieno[3,2-d]pyrimidine-2,4-diamine, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J.A, Green, I, Pollard, H, Howard, T. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 1-Methyl-1H-Imidazole Derivatives as Potent Jak2 Inhibitors.
J.Med.Chem., 57, 2014
|
|
7PNE
 
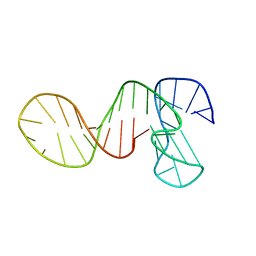 | |
7PNG
 
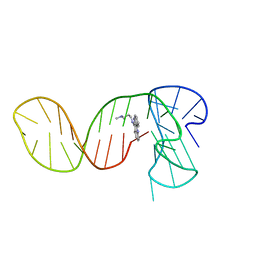 | |
2MCW
 
 | | Solid-state NMR structure of piscidin 3 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
4DH3
 
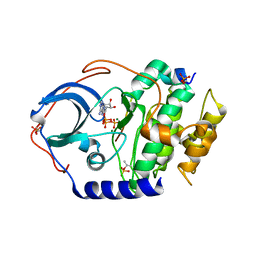 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5HT8
 
 | | Crystal structure of clostrillin double mutant (S17H,S19H) in complex with nickel | | Descriptor: | Beta and gamma crystallin, NICKEL (II) ION | | Authors: | Jamkhindikar, A, Srivastava, S.S, Sankaranarayanan, R. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Transition Metal-Binding, Trimeric beta gamma-Crystallin from Methane-Producing Thermophilic Archaea, Methanosaeta thermophila
Biochemistry, 56, 2017
|
|
4DIK
 
 | | Flavo Di-iron protein H90A mutant from Thermotoga maritima | | Descriptor: | CHLORIDE ION, FLAVOPROTEIN, MU-OXO-DIIRON | | Authors: | Fang, H, Caranto, J.D, Taylor, A.B, Hart, P.J, Kurtz Jr, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Histidine ligand variants of a flavo-diiron protein: effects on structure and activities.
J.Biol.Inorg.Chem., 17, 2012
|
|
4DN4
 
 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
5XTJ
 
 | | Mannanase(RmMan134A) | | Descriptor: | Endo beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Huang, P. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
6EEL
 
 | |
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
1X09
 
 | | Crystal structure of the D26A mutant UPPs in complex with magnesium and isopentenyl pyrophosphate | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
