4Y1T
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
3F5U
 
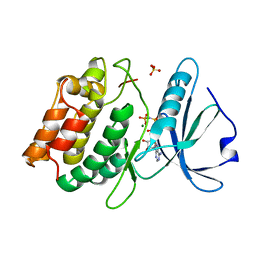 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8F6U
 
 | |
8F6V
 
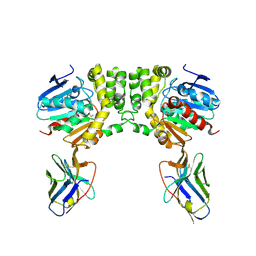 | |
3ZVH
 
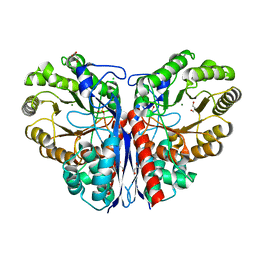 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant Q73A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
5JYC
 
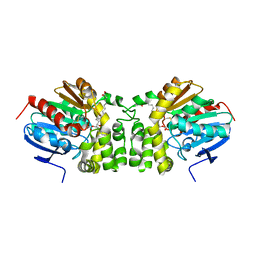 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EET hydrolysis intermediate | | Descriptor: | (5~{Z},11~{Z},14~{R},15~{R})-14,15-bis(oxidanyl)icosa-5,8,11-trienoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8THI
 
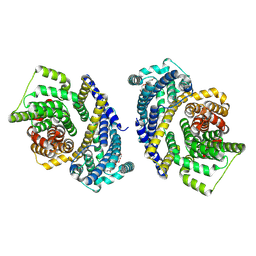 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
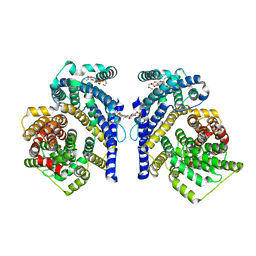 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
3ZVI
 
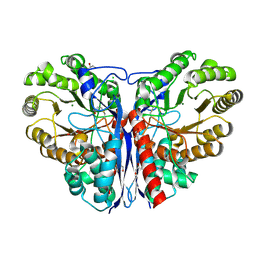 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant L384A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
4YX9
 
 | |
7KPA
 
 | | asymmetric hTNF-alpha | | Descriptor: | 5-(1-{[2-(difluoromethoxy)phenyl]methyl}-2-{[3-(2-oxopyrrolidin-1-yl)phenoxy]methyl}-1H-benzimidazol-6-yl)pyridin-2(1H)-one, Tumor necrosis factor | | Authors: | Fox III, D, Lukacs, C.M, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF.
Nat Commun, 12, 2021
|
|
5M42
 
 | | Structure of Thermus thermophilus L-proline dehydrogenase lacking alpha helices A, B and C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Proline dehydrogenase | | Authors: | Martinez-Julvez, M, Huijbers, M.M.E, van Berkel, W.J.H, Medina, M. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline dehydrogenase from Thermus thermophilus does not discriminate between FAD and FMN as cofactor.
Sci Rep, 7, 2017
|
|
7KP7
 
 | | asymmetric mTNF-alpha hTNFR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor, ... | | Authors: | Arakaki, T.L, Fox III, D, Edwards, T.E, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
6FV6
 
 | | Monomer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Monomer structure of Aq128 in the outward-facing state
To Be Published
|
|
6FV8
 
 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimer structure of the MATE family multidrug resistance transporter Aq128
in the outward-facing state
To Be Published
|
|
6FV7
 
 | | Dimer structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dimer structure of Aq128 in the outward-facing state
To Be Published
|
|
8R2E
 
 | |
8R2B
 
 | |
7EPG
 
 | |
7EPI
 
 | |
7EPJ
 
 | |
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
8R66
 
 | | Crystal structure of ThsA Macro domain in complex with signaling molecule | | Descriptor: | (2~{S})-3-[1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]imidazol-4-yl]-2-azanyl-propanoic acid, SULFATE ION, Thoeris protein ThsA | | Authors: | Tamulaitiene, G, Sabonis, D. | | Deposit date: | 2023-11-21 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | TIR domains produce histidine-ADPR as an immune signal in bacteria
Nature, 2025
|
|
4ZEJ
 
 | |
5BOI
 
 | |
