8SI3
 
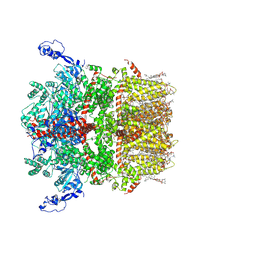 | | Cryo-EM structure of TRPM7 in GDN detergent in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI2
 
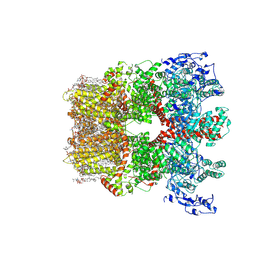 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CHOLESTEROL, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SF0
 
 | | Cryo-EM Structure of RyR1 + cAMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEZ
 
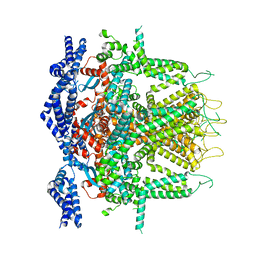 | | Cryo-EM Structure of RyR1 + Adenine (Local Refinement of TMD) | | Descriptor: | ADENINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
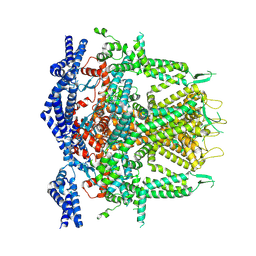 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | Descriptor: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEX
 
 | | Cryo-EM Structure of RyR1 + AMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEW
 
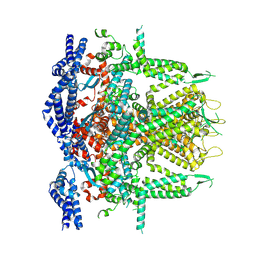 | | Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEU
 
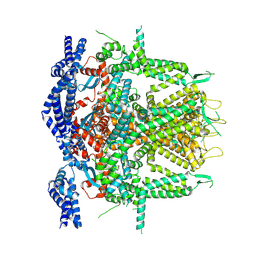 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
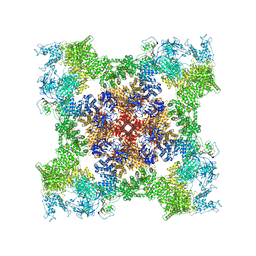 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
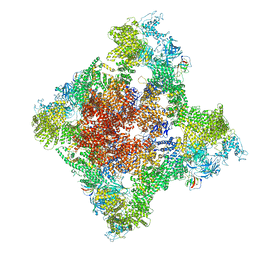 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
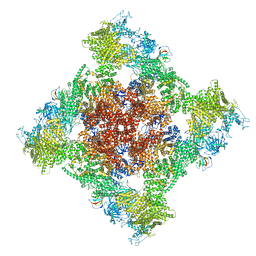 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SDA
 
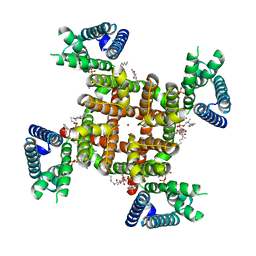 | | CryoEM structure of rat Kv2.1(1-598) L403A mutant in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
8SD3
 
 | | CryoEM structure of rat Kv2.1(1-598) wild type in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
8S9C
 
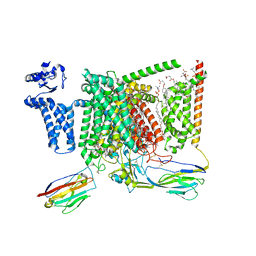 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9B
 
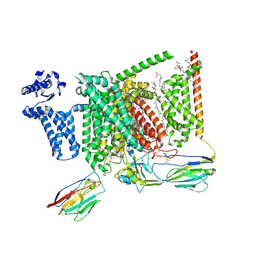 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S3E
 
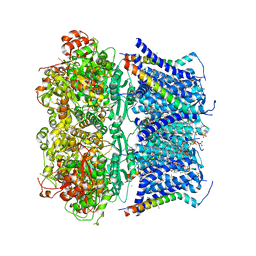 | | Structure of rabbit Slo1 in complex with gamma1/LRRC26 | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Redhardt, M, Raunser, S, Raisch, T. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Cryo-EM structure of the Slo1 potassium channel with the auxiliary gamma 1 subunit suggests a mechanism for depolarization-independent activation.
Febs Lett., 598, 2024
|
|
8QZ4
 
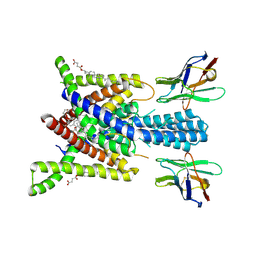 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb76) | | Descriptor: | BARIUM ION, CHOLESTEROL HEMISUCCINATE, Nanobody 76, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ3
 
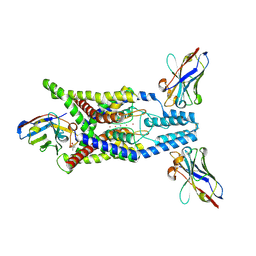 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ2
 
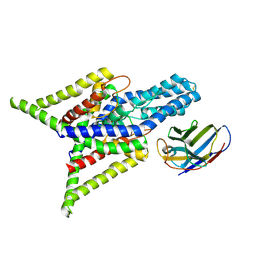 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an inhibitory nanobody (Nb61) | | Descriptor: | Nanobody 61, POTASSIUM ION, Potassium channel subfamily K member 10 | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
