5TY4
 
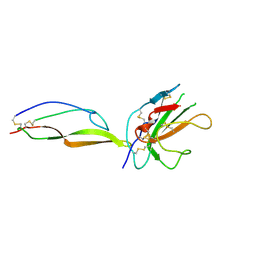 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
8R8M
 
 | |
5U14
 
 | | E. coli dihydropteroate synthase complexed with an 8-mercaptoguanine derivative: 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide | | Descriptor: | 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
8R6C
 
 | |
6CCM
 
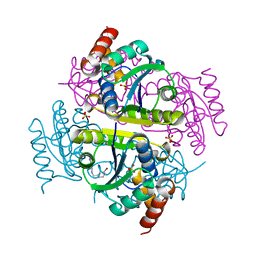 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
 | |
6CHN
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with methyl (R)-4-(3-(2-cyano-1-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)ethyl)phenoxy)piperidine-1-carboxylate | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
5U2N
 
 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
6CAS
 
 | |
8RRP
 
 | |
6CKW
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-3-((7-(((S)-2-amino-2-(2-methoxyphenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)-3-(3-chlorophenyl)propanenitrile | | Descriptor: | (3R)-3-[(7-{[(2S)-2-amino-2-(2-methoxyphenyl)ethyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]-3-(3-chlorophenyl)propanenitrile, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CHO
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-2-((1-(3-(4-methoxyphenoxy)phenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-({(1R)-1-[3-(4-methoxyphenoxy)phenyl]ethyl}amino)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CAR
 
 | | Serial Femtosecond X-ray Crystal Structure of 30S ribosomal subunit from Thermus thermophilus in complex with Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H. | | Deposit date: | 2018-01-31 | | Release date: | 2018-07-25 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature.
Nucleic Acids Res., 46, 2018
|
|
6CCO
 
 | |
6CCQ
 
 | |
8RD5
 
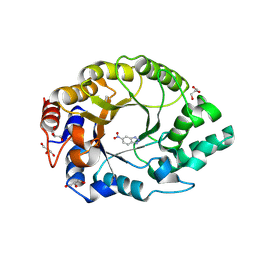 | | Crystal structure of Kemp Eliminase HG3.R5 with bound transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Schaub, D, Schwander, T, Hueppi, S, Buller, R.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enriching productive mutational paths accelerates enzyme evolution.
Nat.Chem.Biol., 2024
|
|
5UIS
 
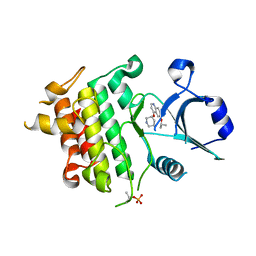 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
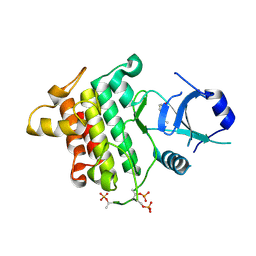 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
6CHM
 
 | |
6CHP
 
 | |
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | Descriptor: | Klebsidin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
6CT5
 
 | |
6CXZ
 
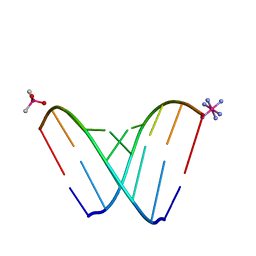 | | RNA octamer containing 2'-F, 4'-Calpha-Me U. | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*AP*AP*(U4M)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
