6W8D
 
 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | Descriptor: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
1IC7
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
6W7M
 
 | | 30S-Inactive-high-Mg2+ + carbon layer | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
7BMC
 
 | | Crystal structure of 14-3-3 sigma in complex with CIP2ApS904 peptide and stabilizing Fusicoccin A | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, FUSICOCCIN, ... | | Authors: | Centorrino, F, Andlovic, B, Ottmann, C. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fusicoccin-A Targets Cancerous Inhibitor of Protein Phosphatase 2A by Stabilizing a C-Terminal Interaction with 14-3-3.
Acs Chem.Biol., 17, 2022
|
|
6WA6
 
 | |
5K8Q
 
 | | Crystal Structure of Calcium-loaded Calmodulin in complex with STRA6 CaMBP2-site peptide. | | Descriptor: | CALCIUM ION, Calmodulin, IMIDAZOLE, ... | | Authors: | Stowe, S.D, Clarke, O.B, Cavalier, M.C, Godoy-Ruiz, R, Mancia, F, Weber, D.J. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
7BM9
 
 | | Crystal structure of 14-3-3 sigma in complex with CIP2ApS904 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Centorrino, F, Andlovic, B, Ottmann, C. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fusicoccin-A Targets Cancerous Inhibitor of Protein Phosphatase 2A by Stabilizing a C-Terminal Interaction with 14-3-3.
Acs Chem.Biol., 17, 2022
|
|
1I2D
 
 | |
6CNO
 
 | |
6CRJ
 
 | |
6TDX
 
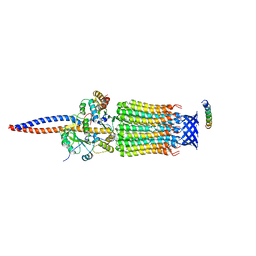 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, rotor, rotational state 1 | | Descriptor: | ATP synthase F1 subunit epsilon, ATP synthase F1 subunit gamma, ATP synthase subunit c, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
1IC5
 
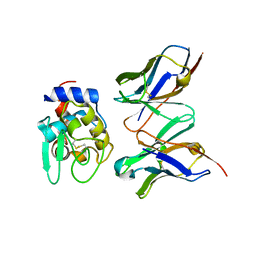 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1I54
 
 | | CYTOCHROME C (TUNA) 2FE:1ZN MIXED-METAL PORPHYRINS | | Descriptor: | CYTOCHROME C, HEME C, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Crane, B.R, Tezcan, F.A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-02-24 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electron tunneling in protein crystals.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KO7
 
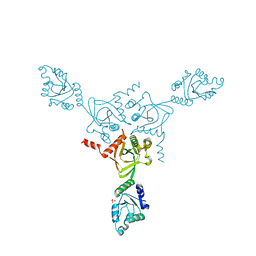 | | X-ray structure of the HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution | | Descriptor: | Hpr kinase/phosphatase, PHOSPHATE ION | | Authors: | Marquez, J.A, Hasenbein, S, Koch, B, Fieulaine, S, Nessler, S, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2K2M
 
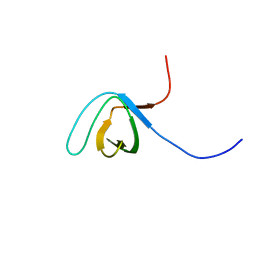 | | Structural Basis of PxxDY Motif Recognition in SH3 Binding | | Descriptor: | Eps8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding.
J.Mol.Biol., 382, 2008
|
|
7XN6
 
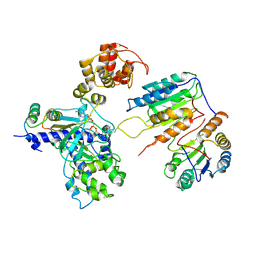 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
6BRM
 
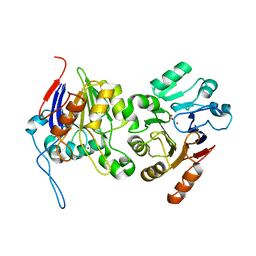 | | The crystal structure of isothiocyanate hydrolase from Delia radicum gut bacteria | | Descriptor: | FORMIC ACID, Putative metal-dependent isothiocyanate hydrolase SaxA, ZINC ION | | Authors: | Tan, K, van den Bosch, T, Joachimiak, A, Welte, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Functional Profiling and Crystal Structures of Isothiocyanate Hydrolases Found in Gut-Associated and Plant-Pathogenic Bacteria.
Appl. Environ. Microbiol., 84, 2018
|
|
6NPC
 
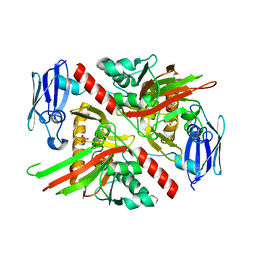 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and 2-trimethylaminoethylphosphonate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, N,N,N-trimethyl-2-phosphonoethan-1-aminium, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
7CA3
 
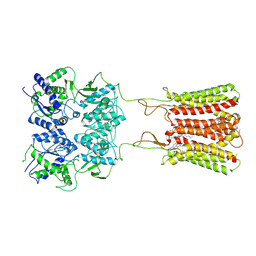 | | Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
2JKZ
 
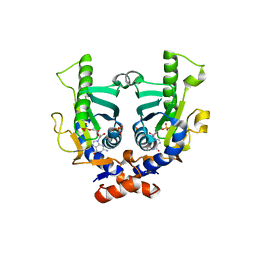 | | SACCHAROMYCES CEREVISIAE HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH GMP (GUANOSINE 5'- MONOPHOSPHATE) (ORTHORHOMBIC CRYSTAL FORM) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, SULFATE ION | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2008-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
2JBG
 
 | |
6TRI
 
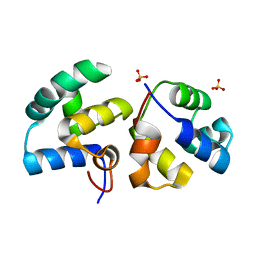 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MDX
 
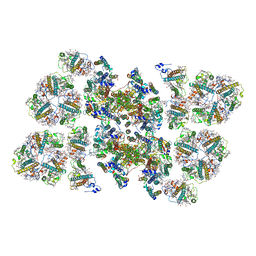 | | Cryo-EM structure of the PSII supercomplex from Arabidopsis thaliana | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein 1, ... | | Authors: | van Bezouwen, L.S, Caffarri, S, Kale, R.S, Kouril, R, Thunnissen, A.M.W.H, Oostergetel, G.T, Boekema, E.J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-06-21 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Subunit and chlorophyll organization of the plant photosystem II supercomplex.
Nat Plants, 3, 2017
|
|
2KC2
 
 | | NMR structure of the F1 domain (residues 86-202) of the talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Elliott, P.R, Roberts, G.C.K, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2008-12-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation.
Embo J., 29, 2010
|
|
