3CCV
 
 | |
7VYH
 
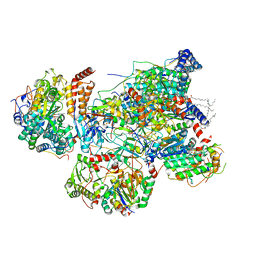 | | Matrix arm of deactive state CI from Rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OTC
 
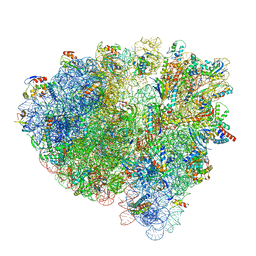 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1GPA
 
 | |
6ORE
 
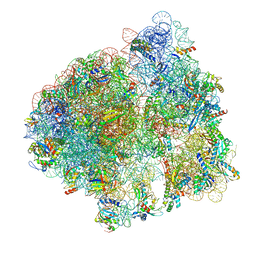 | | Release complex 70S | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
4V9P
 
 | |
7W2K
 
 | | Deactive state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3CCL
 
 | |
6I7V
 
 | | Ribosomal protein paralogs bL31 and bL36 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Pulk, A, Cate, J.H.D, Remme, J, Lilleorg, S, Reier, K, Peil, L, Liiv, A, Tammsalu, T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacterial ribosome heterogeneity: Changes in ribosomal protein composition during transition into stationary growth phase.
Biochimie, 156, 2018
|
|
2QEX
 
 | |
7AS8
 
 | | Bacillus subtilis ribosome quality control complex state B. Ribosomal 50S subunit with P-tRNA, RqcH, and RqcP/YabO | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-09 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for Bacterial Ribosome-Associated Quality Control by RqcH and RqcP.
Mol.Cell, 81, 2021
|
|
3CCQ
 
 | |
6WOO
 
 | | CryoEM structure of yeast 80S ribosome with Met-tRNAiMet, eIF5B, and GDP | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal rRNA, ... | | Authors: | Wang, J, Wang, J, Puglisi, J, Fernandez, I.S. | | Deposit date: | 2020-04-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An active role of the eukaryotic large ribosomal subunit in translation initiation fidelity.
To Be Published
|
|
1YJW
 
 | | Crystal Structure Of Quinupristin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-15 | | Release date: | 2005-04-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
2OTJ
 
 | | 13-deoxytedanolide bound to the large subunit of Haloarcula marismortui | | Descriptor: | 13-DEOXYTEDANOLIDE, 23S ribosomal RNA, 50S ribosomal protein L10E, ... | | Authors: | Blaha, G, Schroeder, S.J, Tirado-Rives, J. | | Deposit date: | 2007-02-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structures of Antibiotics Bound to the E Site Region of the 50 S Ribosomal Subunit of Haloarcula marismortui: 13-Deoxytedanolide and Girodazole.
J.Mol.Biol., 367, 2007
|
|
7UG6
 
 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1E4O
 
 | | Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phosphorylase Recognition and Phosphorylysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
5T2A
 
 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
3I56
 
 | | Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
41BI
 
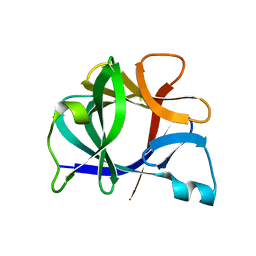 | |
7B5K
 
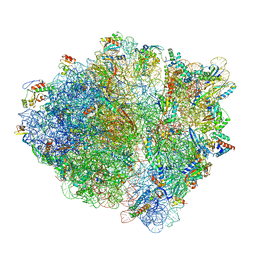 | | E. coli 70S containing suppressor tRNA in the A-site stabilized by a Negamycin analogue and P-site tRNA-nascent chain. | | Descriptor: | 16S rRNA, 2-[[[(3~{R},5~{R})-3-azanyl-6-(3-azanylpropylamino)-5-oxidanyl-hexanoyl]amino]-methyl-amino]ethanoic acid, 23S rRNA, ... | | Authors: | Albers, S, Beckert, B, Matthies, M, Schuster, R, Riedner, M, Seuring, C, Sanyal, S, Torda, A, Wilson, N.D, Ignatova, Z. | | Deposit date: | 2020-12-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Harnessing evolutionarily flexibility in tRNAs to maximize nonsense suppression activities
Nat Commun, 2021
|
|
5AFI
 
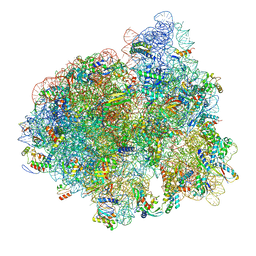 | | 2.9A Structure of E. coli ribosome-EF-TU complex by cs-corrected cryo-EM | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Konevega, A.L, Bock, L.V, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the E. coli ribosome-EF-Tu complex at <3 angstrom resolution by Cs-corrected cryo-EM.
Nature, 520, 2015
|
|
7GPB
 
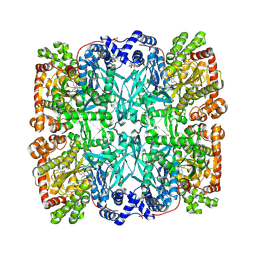 | | STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Barford, D, Hu, S.-H, Johnson, L.N. | | Deposit date: | 1990-11-13 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP.
J.Mol.Biol., 218, 1991
|
|
2XV7
 
 | | Crystal structure of vascular endothelial growth factor D | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR D, alpha-D-mannopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Leppanen, V.-M, Jeltsch, M, Anisimov, A, Tvorogov, D, Aho, K, Kalkkinen, N, Toivanen, P, Yla-Herttuala, S, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-10-23 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Determinants of Vascular Endothelial Growth Factor-D - Receptor Binding and Specificity.
Blood, 117, 2011
|
|
7UNU
 
 | | Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (composite structure I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Basu, R.S, Sherman, M.B, Gagnon, M.G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Nat Commun, 13, 2022
|
|
