7QRD
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
6S01
 
 | | Structure of LEDGF PWWP domain bound H3K36 methylated nucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Wang, H, Farnung, L, Dienemann, C, Cramer, P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding.
Nat.Struct.Mol.Biol., 27, 2020
|
|
9EAR
 
 | | CHD1-nucleosome complex (closed state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1, ... | | Authors: | James, A.M, Farnung, L. | | Deposit date: | 2024-11-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of human CHD1 nucleosome recruitment and pausing.
Mol.Cell, 85, 2025
|
|
6RYU
 
 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
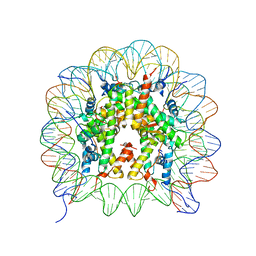
5B40
 
 | | The nucleosome structure containing H2B-K120 and H4-K31 monoubiquitinations | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Machida, S, Sekine, S, Nishiyama, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
To Be Published
|
|
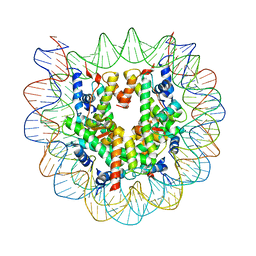
3B6F
 
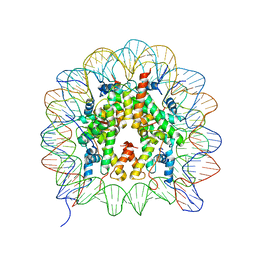 | |
2F8N
 
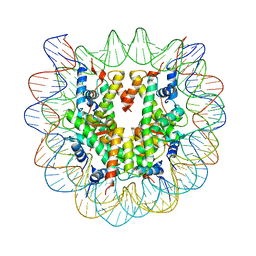 | |
3B6G
 
 | |
2FJ7
 
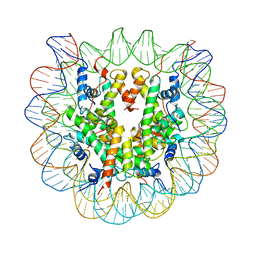 | | Crystal structure of Nucleosome Core Particle Containing a Poly (dA.dT) Sequence Element | | Descriptor: | 147 bp DNA containing 16 bp poly dA element, 147 bp DNA containing 16 bp poly dT element, Histone H2B, ... | | Authors: | Bao, Y, White, C.L, Luger, K. | | Deposit date: | 2005-12-31 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleosome Core Particles Containing a Poly(dA.dT) Sequence Element Exhibit a Locally Distorted DNA Structure.
J.Mol.Biol., 361, 2006
|
|
5NL0
 
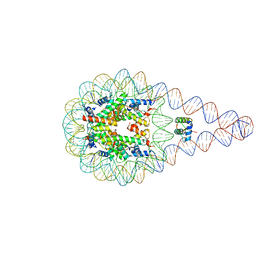 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
5OXV
 
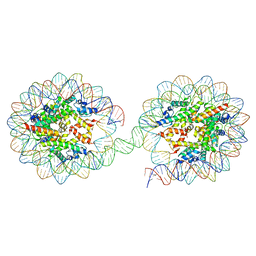 | | Structure of the 4_601_157 tetranucleosome (C2 form) | | Descriptor: | DNA STRAND 1 (601-based sequence model), DNA STRAND 2 (601-based sequence model), Histone H2A, ... | | Authors: | Ekundayo, B, Schalch, T. | | Deposit date: | 2017-09-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.721 Å) | | Cite: | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
5OY7
 
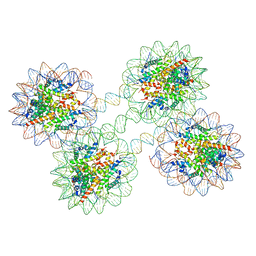 | | Structure of the 4_601_157 tetranucleosome (P1 form) | | Descriptor: | CHLORIDE ION, DNA (619-MER), Histone H2A, ... | | Authors: | Ekundayo, B, Richmond, T.J, Schalch, T. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.774 Å) | | Cite: | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
7OAO
 
 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
6T9L
 
 | |
5X0Y
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5T0K
 
 | | Structure of G9a SET-domain with H3K9M mutant peptide and SAM | | Descriptor: | H3K9 mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading.
Elife, 5, 2016
|
|
2NZD
 
 | | Nucleosome core particle containing 145 bp of DNA | | Descriptor: | DNA (145-MER), Histone H2B, Histone H3, ... | | Authors: | Ong, M.S, Richmond, T.J, Davey, C.A. | | Deposit date: | 2006-11-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA stretching and extreme kinking in the nucleosome core
J.Mol.Biol., 368, 2007
|
|
5T0M
 
 | |
5O9G
 
 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
5XF6
 
 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having an ethylenediamine linker | | Descriptor: | DNA (145-MER), ETHANE-1,2-DIAMINE, Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5X0X
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
2OQ7
 
 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
6UXW
 
 | | SWI/SNF nucleosome complex with ADP-BeFx | | Descriptor: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8.96 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
2KWJ
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
8UT7
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
