6JAX
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
7LK9
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 HC + COVD21-C8 LC | | Descriptor: | COV107-23 heavy chain, COVD21-C8 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LKA
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 | | Descriptor: | ACETATE ION, COV107-23 heavy chain, COV107-23 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
6JKD
 
 | | Crystal structure of tetrameric PepTSo2 in I4 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NM8
 
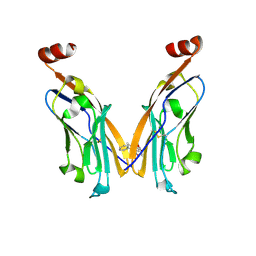 | | IgV-V76T BMS compound 105 | | Descriptor: | N-({2,6-dimethoxy-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)-D-alanine, Programmed cell death 1 ligand 1 | | Authors: | Perry, E, Zhao, B, Fesik, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6NNJ
 
 | |
6NV6
 
 | |
7LG6
 
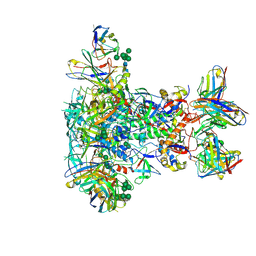 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
6NNF
 
 | |
7LUS
 
 | | IgG2 Fc Charge Pair Mutation version 1 (CPMv1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 2 | | Authors: | Sudom, A, Whittington, D, Garces, F, Wang, Z. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Fc scaffold for multispecific antibodies.
Iscience, 24, 2021
|
|
6JKC
 
 | | Crystal structure of tetrameric PepTSo2 in P4212 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NF2
 
 | |
6NP9
 
 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6NM6
 
 | |
6NNV
 
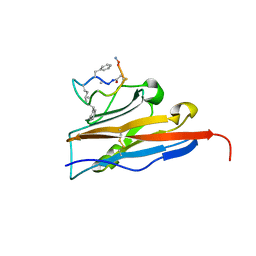 | | PD-L1 IgV domain complex with macro-cyclic peptide | | Descriptor: | Programmed cell death 1 ligand 1, macrocyclic peptide | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7LX2
 
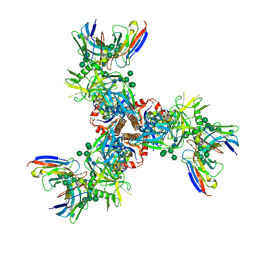 | | Cryo-EM structure of ConSOSL.UFO.664 (ConS) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
7LX3
 
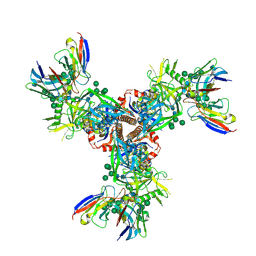 | | Cryo-EM structure of EDC-crosslinked ConSOSL.UFO.664 (ConS-EDC) in complex with bNAb PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Env glycoprotein gp160, ... | | Authors: | Martin, G.M, Ward, A.B, Sattentau, Q.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Npj Vaccines, 8, 2023
|
|
6NOJ
 
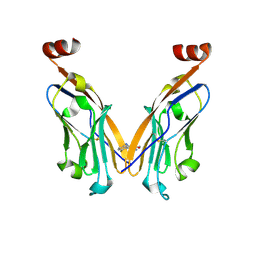 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | Programmed cell death 1 ligand 1, methyl 3-amino-4-(2-fluorophenyl)-1H-pyrrole-2-carboxylate | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6KA7
 
 | | The complex structure of Human IgG Fc and its binding Repebody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, repebody | | Authors: | Choi, J, Kim, H. | | Deposit date: | 2019-06-21 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The complex structure of Human IgG Fc and its binding Repebody
To Be Published
|
|
5HBV
 
 | | Complex structure of Fab35 and mouse nAChR alpha1 | | Descriptor: | Acetylcholine receptor subunit alpha 1, Alpha-bungarotoxin isoform V31, Fab35, ... | | Authors: | Noridomi, K, Watanabe, G, Hansen, M.N, Han, G.W, Chen, L. | | Deposit date: | 2016-01-02 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanisms of myasthenia gravis and their therapeutic implications.
Elife, 6, 2017
|
|
6NM7
 
 | | PD-L1 IgV domain bound to fragment | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, Programmed cell death 1 ligand 1 | | Authors: | Perry, E, Zhao, B. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6NXV
 
 | | Crystal structure of the theta class glutathione S-transferase from the citrus canker pathogen Xanthomonas axonopodis pv. citri, apo form | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Glutathione S-transferase | | Authors: | Hilario, E, De Keyser, S, Fan, L. | | Deposit date: | 2019-02-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical characterization of a glutathione transferase from the citrus canker pathogen Xanthomonas.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5HYE
 
 | | Glycosylated Knob-Knob Fc fragment (P212121) | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products.
Protein Eng. Des. Sel., 30, 2017
|
|
5HYF
 
 | | Glycosylated Knob-Knob Fc fragment (P6122) | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products.
Protein Eng. Des. Sel., 30, 2017
|
|
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
