3BLM
 
 | |
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
3TWR
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human 3BP2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SH3 domain-binding protein 2, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3U9Y
 
 | |
3TII
 
 | | Tubulin tyrosine ligase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ttl protein | | Authors: | Roll-Mecak, A, Szyk, A, Deaconescu, A, Piszczek, G. | | Deposit date: | 2011-08-20 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1EEG
 
 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
3ORZ
 
 | | PDK1 mutant bound to allosteric disulfide fragment activator 2A2 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(3-chlorophenyl)piperazin-1-yl]-4-oxobutane-1-thiol | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-08 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9995 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P0Q
 
 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | N-[2-(4-chlorophenyl)ethyl]-6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-amine, SODIUM ION, SULFATE ION, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
3TWV
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human NUMA1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
3QH6
 
 | | 1.8A resolution structure of CT296 from Chlamydia trachomatis | | Descriptor: | CT296, TETRAETHYLENE GLYCOL | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J.Bacteriol., 193, 2011
|
|
3QHT
 
 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1M7Q
 
 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3PG3
 
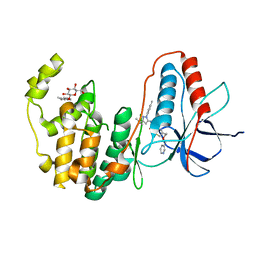 | | Human p38 MAP Kinase in Complex with RL182 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{4-[2-(pyridin-3-ylmethoxy)ethyl]-1,3-thiazol-2-yl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of cell active N-pyrazole, N-thiazole urea inhibitors of the MAP kinase p38alpha
To be Published
|
|
1MD9
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB AND AMP | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3TW2
 
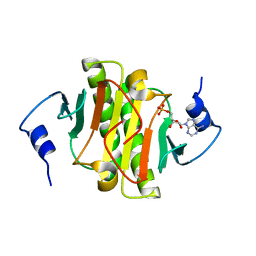 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3Q48
 
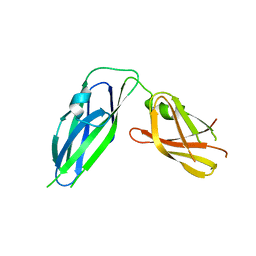 | | Crystal structure of Pseudomonas aeruginosa CupB2 chaperone | | Descriptor: | Chaperone CupB2 | | Authors: | Cai, X, Wang, R, Filloux, A, Waksman, G, Meng, G. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones
Plos One, 6, 2011
|
|
3U23
 
 | | Atomic resolution crystal structure of the 2nd SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide from human RIN3 | | Descriptor: | 1,2-ETHANEDIOL, CD2-associated protein, Ras and Rab interactor 3 | | Authors: | Simister, P.C, Rouka, E, Janning, M, Muniz, J.R.C, Kirsch, K.H, Knapp, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Krojer, T, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
1H2O
 
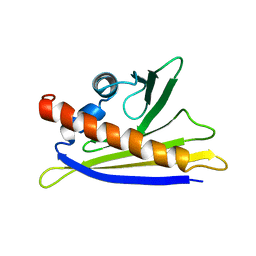 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
1VHQ
 
 | |
3TWX
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human FNBP1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3UA9
 
 | | Crystal structure of human tankyrase 2 in complex with a selective inhibitor | | Descriptor: | 4-[(3aR,4S,7R,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Narwal, M, Lehtio, L. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of selective inhibition of human tankyrases.
J.Med.Chem., 55, 2012
|
|
3U5P
 
 | |
3TWT
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human MCL1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, NONAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TZS
 
 | |
