2MEY
 
 | |
1X67
 
 | | Solution structure of the cofilin homology domain of HIP-55 (drebrin-like protein) | | Descriptor: | Drebrin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
2F40
 
 | |
2MV4
 
 | |
2ETZ
 
 | | The NMR minimized average structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
2EU0
 
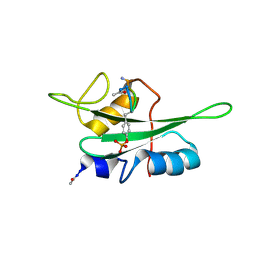 | | The NMR ensemble structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
2NCI
 
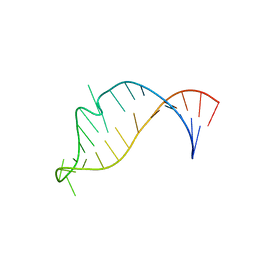 | |
2M99
 
 | |
2MZV
 
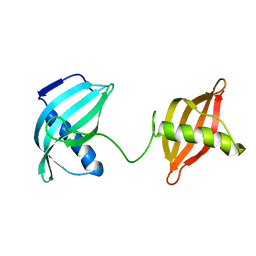 | |
2NR1
 
 | | TRANSMEMBRANE SEGMENT 2 OF NMDA RECEPTOR NR1, NMR, 10 STRUCTURES | | Descriptor: | NR1 M2 | | Authors: | Gesell, J.J, Sun, W, Montal, M, Opella, S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
2MHW
 
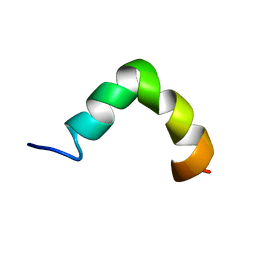 | | The solution NMR structure of maximin-4 in SDS micelles | | Descriptor: | Antimicrobial peptide | | Authors: | Toke, O, Banoczi, Z, Kiraly, P, Heinzmann, R, Burck, J, Ulrich, A.S, Hudecz, F. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A kinked antimicrobial peptide from Bombina maxima. I. Three-dimensional structure determined by NMR in membrane-mimicking environments.
Eur.Biophys.J., 40, 2011
|
|
2NBI
 
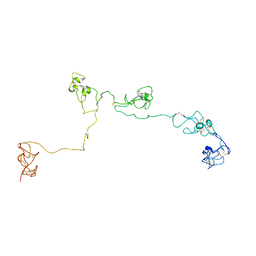 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
2LJW
 
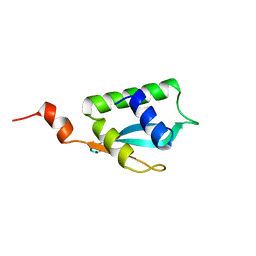 | | Solution NMR structure of Alr2454 protein from Nostoc sp. strain PCC 7120, Northeast Structural Genomics Consortium Target NsR264 | | Descriptor: | Alr2454 protein | | Authors: | Aramini, J.M, Lee, D, Ciccosanti, C, Janjua, H, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Alr2454 from Nostoc sp. PCC 7120, the first structural representative of Pfam domain family PF11267.
J.Struct.Funct.Genom., 13, 2012
|
|
2M3G
 
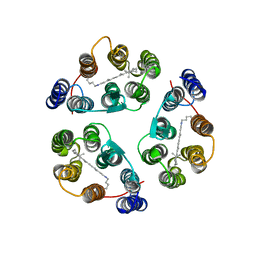 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy | | Descriptor: | Anabaena Sensory Rhodopsin, RETINAL | | Authors: | Wang, S, Munro, R.A, Shi, L, Kawamura, I, Okitsu, T, Wada, A, Kim, S, Jung, K, Brown, L.S, Ladizhansky, V. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein.
Nat.Methods, 10, 2013
|
|
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
1XC0
 
 | | Twenty Lowest Energy Structures of Pa4 by Solution NMR | | Descriptor: | Pardaxin P-4 | | Authors: | Porcelli, F, Buck, B, Lee, D.-K, Hallock, K.J, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of pardaxin determined by NMR experiments in model membranes
J.Biol.Chem., 279, 2004
|
|
2MEM
 
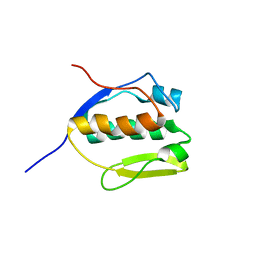 | | Solution NMR structure of SLED domain of Scml2 | | Descriptor: | Sex comb on midleg-like protein 2 | | Authors: | Bezsonova, I. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-binding Domain from Scml2 (Sex Comb on Midleg-like 2).
J.Biol.Chem., 289, 2014
|
|
2O8Z
 
 | | Bound Structure of CRF1 Extracellular Domain Antagonist | | Descriptor: | cCRF(30-41) Peptide | | Authors: | Mesleh, M.F, Shirley, W.A, Heise, C.E, Ling, N, Maki, R.A, Laura, R.P. | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization of a minimal peptide antagonist bound to the extracellular domain of the corticotropin-releasing factor1 receptor.
J.Biol.Chem., 282, 2007
|
|
1RGD
 
 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1UUU
 
 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
2OV6
 
 | |
1JHB
 
 | |
1SPF
 
 | |
1TCH
 
 | |
