3J0Q
 
 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
4XUI
 
 | |
4D88
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXQ490 | | Descriptor: | (3S,4S,5R)-3-{4-amino-3-fluoro-5-[(2S)-3,3,3-trifluoro-2-hydroxypropyl]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
4D85
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BVI151 | | Descriptor: | (3R,4S,5S)-3-[(3-tert-butylbenzyl)amino]-5-[(4,4,7'-trifluoro-1',2'-dihydrospiro[cyclohexane-1,3'-indol]-5'-yl)methyl]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, IODIDE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
3J78
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class I - non-rotated ribosome with 2 tRNAs) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3J77
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
4R3Z
 
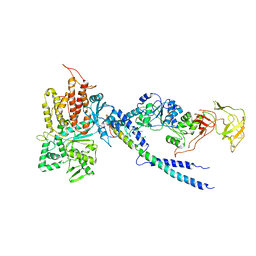 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZOU
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, DIMETHYL SULFOXIDE, FARNESYL PYROPHOSPHATE SYNTHASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZJ3
 
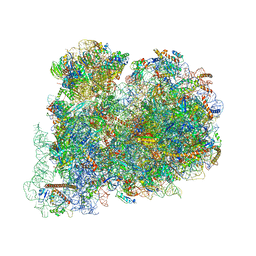 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
6Z6K
 
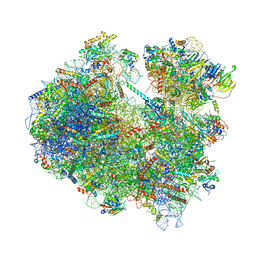 | | Cryo-EM structure of yeast reconstituted Lso2 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6THO
 
 | |
4ACG
 
 | | GSK3b in complex with inhibitor | | Descriptor: | 2-AMINO-5-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-N-[4-(PYRROLIDIN-1-YLMETHYL)PYRIDIN-3-YL]PYRIDINE-3-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
1R57
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
6GT7
 
 | |
3HW2
 
 | | Crystal structure of the SifA-SKIP(PH) complex | | Descriptor: | Pleckstrin homology domain-containing family M member 2, Protein sifA | | Authors: | Diacovich, L, Dumont, A, Lafitte, D, Soprano, E, Guilhon, A.-A, Bignon, C, Gorvel, J.-P, Bourne, Y, Meresse, S. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Interaction between the SifA virulence factor and its host target skip is essential for salmonella pathogenesis
J.Biol.Chem., 284, 2009
|
|
4BVX
 
 | | Crystal structure of the AIMP3-MRS N-terminal domain complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, METHIONINE--TRNA LIGASE, ... | | Authors: | Cho, H.Y, Seo, W.W, Cho, H.J, Kang, B.S. | | Deposit date: | 2013-06-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
2PGD
 
 | |
6KF9
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
4ACC
 
 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-6-(4-{[2-(DIMETHYLAMINO)ETHYL]SULFAMOYL}PHENYL)-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, DIMETHYL SULFOXIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
7RTT
 
 | | Cryo-EM structure of a TTYH2 cis-dimer | | Descriptor: | CALCIUM ION, Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7RTU
 
 | | Cryo-EM structure of a TTYH2 trans-dimer | | Descriptor: | Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7RTW
 
 | | Cryo-EM structure of a TTYH3 cis-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
4AZE
 
 | | Human DYRK1A in complex with Leucettine L41 | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)-2-(phenylamino)-4H-imidazol-4-one, DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION-REGULATED KINASE 1A | | Authors: | Elkins, J.M, Soundararajan, M, Muniz, J.R.C, Tahtouh, T, Burgy, G, Durieu, E, Lozach, O, Cochet, C, Schmid, R.S, Lo, D.C, Delhommel, F, Carreaux, F, Bazureau, J.P, Meijer, L, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Selectivity, Co-Crystal Structures and Neuroprotective Properties of Leucettines, a Family of Protein Kinase Inhibitors Derived from the Marine Sponge Alkaloid Leucettamine B.
J.Med.Chem., 55, 2012
|
|
7RTV
 
 | | Cryo-EM structure of monomeric TTYH2 | | Descriptor: | Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7UXA
 
 | | Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG | | Descriptor: | MAGNESIUM ION, RNA (78-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Stanley, R.E, Hayne, C.K. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
