5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
5KLE
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose | | Descriptor: | Carbohydrate binding module E1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
7U2J
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, peptidyl P-site fMAC-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
1C0B
 
 | |
5M7K
 
 | | Blastochloris viridis photosynthetic reaction center - RC_vir_xfel | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
6WFG
 
 | | Crystal structure of human Naa50 in complex with an inhibitor (compound 3) identified using DNA encoded library technology | | Descriptor: | (2S)-N-[(2S)-3-[1-(3-tert-butyl-1-methyl-1H-pyrazole-5-carbonyl)piperidin-4-yl]-1-(methylamino)-1-oxopropan-2-yl]-6-oxopiperidine-2-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
1BXW
 
 | |
1C0C
 
 | |
6KKW
 
 | |
2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | Descriptor: | TENGPIN | | Authors: | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
1TYN
 
 | | ATOMIC STRUCTURE OF THE TRYPSIN-CYCLOTHEONAMIDE A COMPLEX: LESSONS FOR THE DESIGN OF SERINE PROTEASE INHIBITORS | | Descriptor: | BETA-TRYPSIN, CYCLOTHEONAMIDE A | | Authors: | Lee, A.Y, Hagihara, M, Karmacharya, R, Albers, M.W, Schreiber, S.L, Clardy, J. | | Deposit date: | 1994-09-19 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Structure of the Trypsin-Cyclotheonamide a Complex: Lessons for the Design of Serine Protease Inhibitors
J.Am.Chem.Soc., 115, 1993
|
|
6D0N
 
 | | Crystal structure of a CLC-type fluoride/proton antiporter, V319G mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5UCG
 
 | | Structure of the PP2C Phosphatase Domain and a Fragment of the Regulatory Domain of the Cell Fate Determinant SpoIIE from Bacillus Subtilis | | Descriptor: | Stage II sporulation protein E | | Authors: | Bradshaw, N, Levdikov, V, Zimanyi, C, Gaudet, R, Wilkinson, A, Losick, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.906 Å) | | Cite: | A widespread family of serine/threonine protein phosphatases shares a common regulatory switch with proteasomal proteases.
Elife, 6, 2017
|
|
6QVZ
 
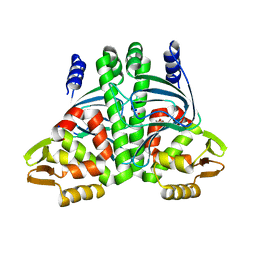 | | The Transcriptional Regulator PrfA-L140H mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6CPE
 
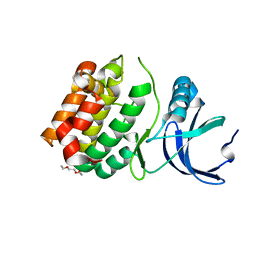 | | Structure of apo, dephosphorylated Aurora A (122-403) in an active conformation | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Aurora kinase A | | Authors: | Otten, R, Kutter, S, Buosi, V, Padua, R.A.P, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
8RRM
 
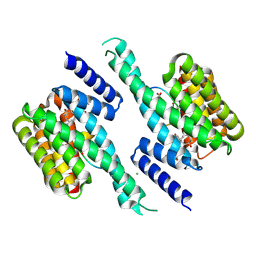 | | tripartite complex between 14-3-3 sigma, Fusicoccin-A, and a phosphopeptide optimized for a Fusicoccin-mediated stabilization of the complex | | Descriptor: | 14-3-3 protein sigma, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Cousido-Siah, A, McEwen, A.G, Monsellier, E, Trave, G. | | Deposit date: | 2024-01-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Holdup Multiplex Assay for High-Throughput Measurement of Protein-Ligand Affinity Constants Using a Mass Spectrometry Readout.
J.Am.Chem.Soc., 147, 2025
|
|
8RRL
 
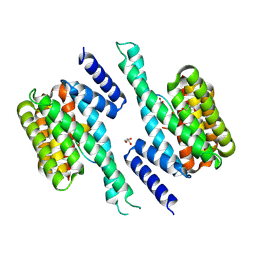 | | 14-3-3 sigma complexed with a phosphopeptide optimized for a Fusicoccin-mediated stabilization of the complex | | Descriptor: | 14-3-3 protein sigma, BICARBONATE ION, GLYCEROL, ... | | Authors: | Cousido-Siah, A, McEwen, A.G, Trave, G, Monsellier, E. | | Deposit date: | 2024-01-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Holdup Multiplex Assay for High-Throughput Measurement of Protein-Ligand Affinity Constants Using a Mass Spectrometry Readout.
J.Am.Chem.Soc., 147, 2025
|
|
8G3R
 
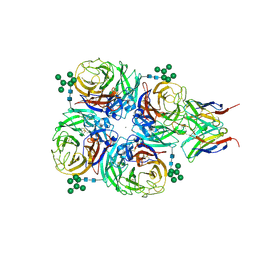 | |
8G3O
 
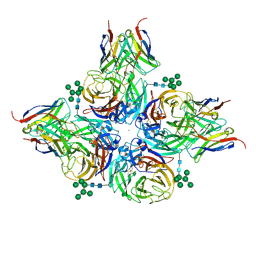 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G40
 
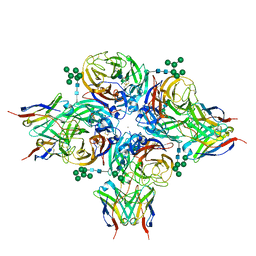 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI19 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G3M
 
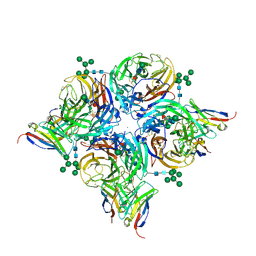 | |
8G3N
 
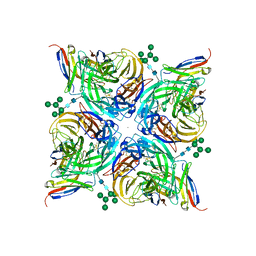 | | N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G3P
 
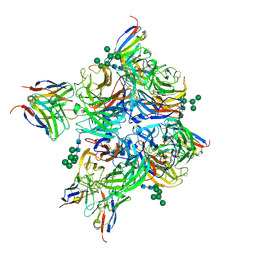 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
5DUI
 
 | |
5E2Y
 
 | |
