6JWJ
 
 | | Npl4 in complex with Ufd1 | | Descriptor: | GLYCEROL, Nuclear protein localization protein 4, Peptide from Ubiquitin fusion degradation protein 1, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
4KEV
 
 | | Crystal structure of SsoPox W263L | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (II) ION | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
6JWT
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613T triple mutant with Pteroate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JX6
 
 | | Tetrameric form of Smac | | Descriptor: | Diablo homolog, mitochondrial | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6KH1
 
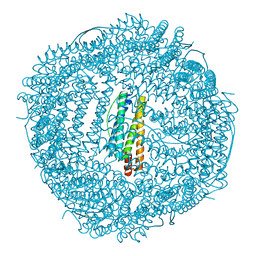 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Whang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
4KF6
 
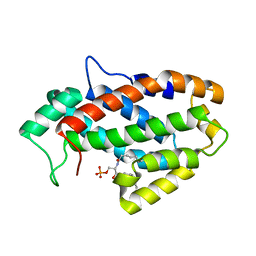 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
6JXI
 
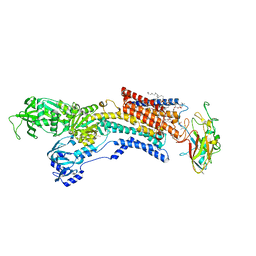 | | Rb+-bound E2-MgF state of the gastric proton pump (Tyr799Trp) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Irie, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single K + -binding site in the crystal structure of the gastric proton pump.
Elife, 8, 2019
|
|
6JYL
 
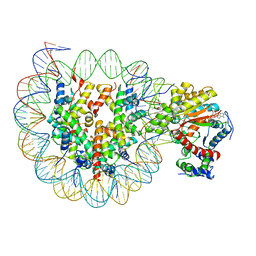 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6KI7
 
 | | Pyrophosphatase mutant K30R from Acinetobacter baumannii | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
3DD2
 
 | | Crystal structure of an RNA aptamer bound to human thrombin | | Descriptor: | ACETIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Long, S.B, Sullenger, B.A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an RNA aptamer bound to thrombin.
Rna, 14, 2008
|
|
5SUU
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17-36. X-ray diffractometer data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION, IODIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
6KIM
 
 | |
6KIR
 
 | | High resolution structure of mouse CXorf40A | | Descriptor: | SULFATE ION, Uncharacterized protein CXorf40 homolog | | Authors: | Wu, B.X. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High resolution structure of mouse CXorf40A
To Be Published
|
|
6KJC
 
 | | lovastatin esterase PcEST, wild type | | Descriptor: | FORMIC ACID, Pc15g00720 protein | | Authors: | Liang, Y.J, Lu, X.F. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural insights into the catalytic mechanism of lovastatin hydrolase
J.Biol.Chem., 2019
|
|
5SUQ
 
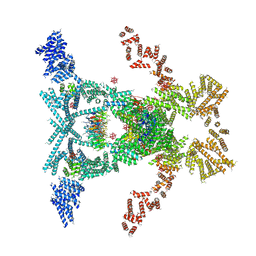 | | Crystal structure of the THO-Sub2 complex | | Descriptor: | 12-TUNGSTOPHOSPHATE, ATP-dependent RNA helicase SUB2, Tex1, ... | | Authors: | Ren, Y, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
6JZC
 
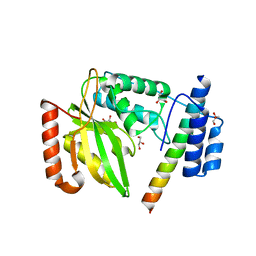 | | Structural basis of tubulin detyrosination | | Descriptor: | GLYCEROL, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6JZF
 
 | |
6JZN
 
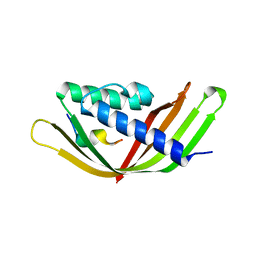 | | Structure of the intermembrane space region of PARC6-PDV1 | | Descriptor: | Peptide from Plastid division protein PDV1, Plastid division protein CDP1, chloroplastic | | Authors: | Feng, Y, Liu, Z. | | Deposit date: | 2019-05-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of PARC6 and PDV1 complex from Arabidopsis thaliana
To Be Published
|
|
6KJH
 
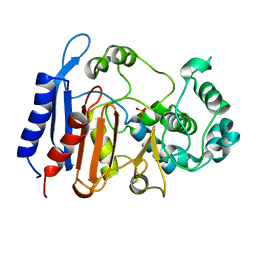 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Sheng, D, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJQ
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJZ
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 1 | | Descriptor: | 5,9-dihydroxy-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H,6H-pyrano[3,2-b]xanthen-6-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
6KK8
 
 | | XN joint refinement of manganese catalase from Thermus Thermophilus HB27 | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (III) ION, OXYGEN ATOM, ... | | Authors: | Yamada, T, Yano, N, Kusaka, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.37 Å), X-RAY DIFFRACTION | | Cite: | Single-crystal time-of-flight neutron Laue methods: application to manganese catalase from Thermus thermophilus HB27
J.Appl.Crystallogr., 2019
|
|
6JZV
 
 | | Crystal structure of SufU from Bacillus subtilis | | Descriptor: | ZINC ION, Zinc-dependent sulfurtransferase SufU | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteine-Persulfide Sulfane Sulfur-Ligated Zn Complex of Sulfur-Carrying SufU in the SufCDSUB System for Fe-S Cluster Biosynthesis.
Inorg.Chem., 2024
|
|
6K08
 
 | | Crystal structure of REV7(R124A/A135D) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
6K0G
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
