3ZW7
 
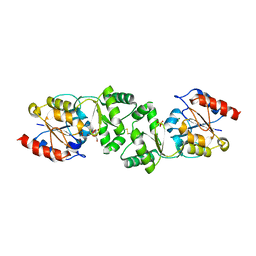 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate and metaphosphate. | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZTY
 
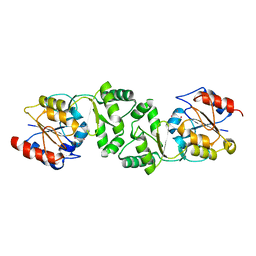 | | The 3-dimensional structure of the gadolinium derivative of MpgP, the mannosyl-3-phosphoglycerate phosphatase from Thermus thermophilus HB27 | | Descriptor: | CHLORIDE ION, GADOLINIUM ATOM, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZX4
 
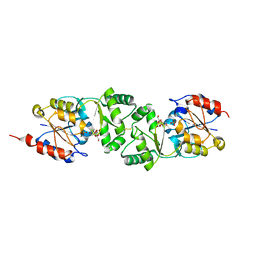 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate,orthophosphate and magnesium | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZX5
 
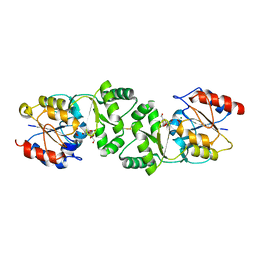 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, covalently bound to vanadate and in complex with alpha- mannosylglycerate and magnesium | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
2Q1C
 
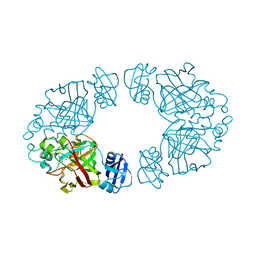 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with calcium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, CALCIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q19
 
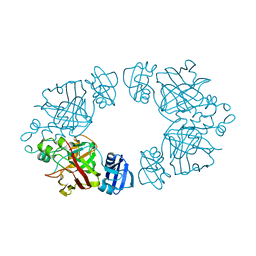 | | 2-keto-3-deoxy-D-arabinonate dehydratase apo form | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q18
 
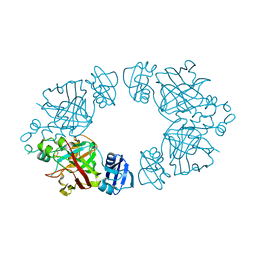 | | 2-keto-3-deoxy-D-arabinonate dehydratase | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase, PHOSPHATE ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q1A
 
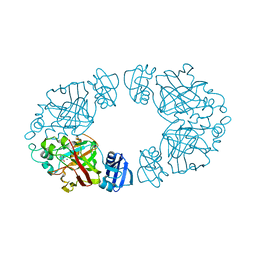 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with magnesium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, MAGNESIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
1MRP
 
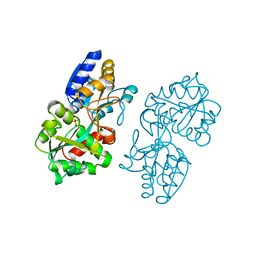 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | Authors: | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | Deposit date: | 1997-05-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
2FCB
 
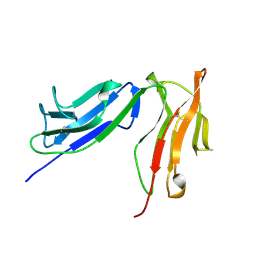 | | HUMAN FC GAMMA RECEPTOR IIB ECTODOMAIN (CD32) | | Descriptor: | PROTEIN (FC GAMMA RIIB) | | Authors: | Sondermann, P, Huber, R, Jacob, U. | | Deposit date: | 1999-01-07 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution.
EMBO J., 18, 1999
|
|
3DJB
 
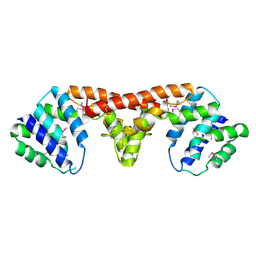 | | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114 | | Descriptor: | Hydrolase, HD family, MAGNESIUM ION | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Vorobiev, S.M, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Maglaqui, M, Owen, L.A, Wang, D, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114
To be Published
|
|
3E29
 
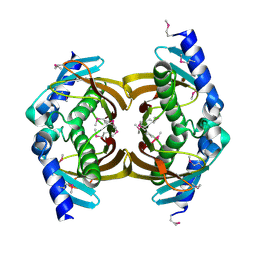 | | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A. | | Descriptor: | uncharacterized protein Q7WE92_BORBR | | Authors: | Kuzin, A.P, Chen, Y, Setharaman, J, Wang, D, Mao, L, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q7WE92_BORBR from thioesterase superfamily. Northeast Structural Genomics Consortium Target BoR214A.
To be Published
|
|
3C7X
 
 | | Hemopexin-like domain of matrix metalloproteinase 14 | | Descriptor: | CHLORIDE ION, Matrix metalloproteinase-14, SODIUM ION | | Authors: | Tochowicz, A, Itoh, Y, Maskos, K, Bode, W, Goettig, P. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions
J.Biol.Chem., 286, 2011
|
|
3E8P
 
 | | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246. | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Janjua, H, Ciccosanti, C, Foote, E.L, Wang, H, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246.
To be Published
|
|
5EKY
 
 | | Crystal structure of deoxyribose-phosphate aldolase from Escherichia coli (K58E-Y96W mutant) | | Descriptor: | 1,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Deoxyribose-phosphate aldolase | | Authors: | Classen, T, Dick, M, Pietruszka, J, Weiergraeber, O.H. | | Deposit date: | 2015-11-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies.
Chem Sci, 7, 2016
|
|
3CRG
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5EL1
 
 | |
5EMU
 
 | |
3EA1
 
 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
4WWI
 
 | |
7AK9
 
 | | Structure of Salmonella TacT3 toxin bound to TacA3 antitoxin C-terminal peptide | | Descriptor: | ABC transporter, Acetyltransferase, DEPHOSPHO COENZYME A | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK7
 
 | | Structure of Salmonella TacT2 toxin bound to TacA2 antitoxin | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CHLORIDE ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK8
 
 | | Structure of Salmonella TacT1 toxin bound to TacA1 antitoxin C-terminal peptide | | Descriptor: | ACETYL COENZYME *A, GCN5 family acetyltransferase, GLYCEROL, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S, Hare, S.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
6BNO
 
 | | Structure of bare actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gurel, P.S, Alushin, G.A. | | Deposit date: | 2017-11-17 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Elife, 6, 2017
|
|
6BNQ
 
 | |
