6W9W
 
 | | R80A PCNA mutant defective in BIR | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Ripley, B.M, Washington, M.T. | | Deposit date: | 2020-03-23 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of PCNA mutant proteins defective in BIR, FF248-249AA and R80A
To be Published
|
|
6WAC
 
 | |
6FZK
 
 | | NMR structure of UB2H, regulatory domain of PBP1b from E. coli | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Egan, A.J.F, Vollmer, W. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
5ZUT
 
 | | Crystal Structure of Yeast PCNA in Complex with N24 Peptide | | Descriptor: | N24, Proliferating cell nuclear antigen | | Authors: | Cheng, X.Y, Kuang, X.L, Zhou, Y, Xia, X.M, SU, Z.D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structure of Yeast PCNA in Complex with N24 Peptide
To Be Published
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
117E
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
4L60
 
 | |
7AOP
 
 | | Structure of NUDT15 in complex with inhibitor TH8321 | | Descriptor: | 2-azanyl-9-cyclohexyl-8-(2-methoxyphenyl)-3~{H}-purine-6-thione, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
7AOM
 
 | | Structure of NUDT15 in complex with Ganciclovir triphosphate | | Descriptor: | Ganciclovir triphosphate, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
4L6P
 
 | |
2HN8
 
 | | Structural characterization and oligomerization of PB1-F2, a pro-apoptotic influenza A virus protein | | Descriptor: | Protein PB1-F2 | | Authors: | Bruns, K, Studtrucker, N, Sharma, A, Fossen, T, Mitzner, D, Eissmann, A, Tessmer, U, Roder, R, Henklein, P, Wray, V, Schubert, U. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterization and oligomerization of PB1-F2, a pro-apoptotic influenza A virus protein.
J.Biol.Chem., 282, 2007
|
|
3JZ0
 
 | | LinB complexed with clindamycin and AMPCPP | | Descriptor: | CLINDAMYCIN, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Lincosamide nucleotidyltransferase, ... | | Authors: | Morar, M, Wright, G.D. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB.
Structure, 17, 2009
|
|
3JYY
 
 | | SeMet LinB complexed with PPi | | Descriptor: | Lincosamide nucleotidyltransferase, MAGNESIUM ION, PYROPHOSPHATE | | Authors: | Morar, M, Wright, G.D. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB.
Structure, 17, 2009
|
|
3KR8
 
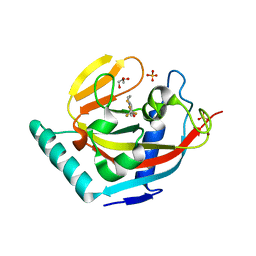 | | Human tankyrase 2 - catalytic PARP domain in complex with an inhibitor XAV939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor.
J.Med.Chem., 53, 2010
|
|
8PRK
 
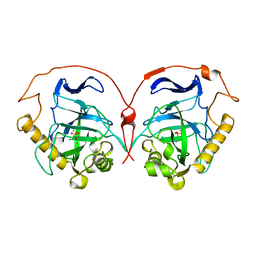 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
4ITD
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*(C6G)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
2KWQ
 
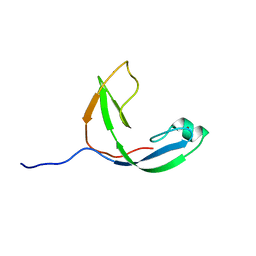 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
4IJ0
 
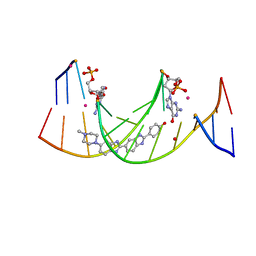 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
1Y7X
 
 | | Solution structure of a two-repeat fragment of major vault protein | | Descriptor: | Major vault protein | | Authors: | Kozlov, G, Vavelyuk, O, Minailiuc, O, Banville, D, Gehring, K, Ekiel, I. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-repeat fragment of major vault protein.
J.Mol.Biol., 356, 2006
|
|
3FWL
 
 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OD8
 
 | | Structure of a peptide derived from Cdc9 bound to PCNA | | Descriptor: | DNA ligase I, mitochondrial precursor, Proliferating cell nuclear antigen | | Authors: | Chapados, B.R, Tainer, J.A. | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase.
Nucleic Acids Res., 35, 2007
|
|
4ESV
 
 | | A New Twist on the Translocation Mechanism of Helicases from the Structure of DnaB with its Substrates | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Itsathitphaisarn, O, Wing, R.A, Eliason, W.K, Wang, J, Steitz, T.A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation.
Cell(Cambridge,Mass.), 151, 2012
|
|
