1CWI
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 2-VAL 3-(N-METHYL)-D-ALANINE CYCLOSPORIN | | Descriptor: | CYCLOSPORIN D, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-25 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
2DW6
 
 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | Descriptor: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ANV
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (2,3-DAP complex) | | Descriptor: | 3-amino-D-alanine, CHLORIDE ION, D-serine dehydratase, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
2FB9
 
 | |
2I57
 
 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
3TRV
 
 | | Crystal structure of quasiracemic villin headpiece subdomain containing (F5Phe17) substitution | | Descriptor: | D-Villin-1, ISOPROPYL ALCOHOL, L-Villin-1, ... | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
2JUE
 
 | |
2DNS
 
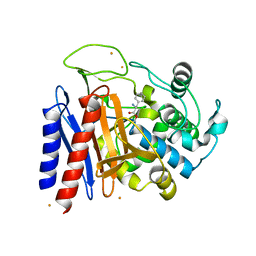 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2KQL
 
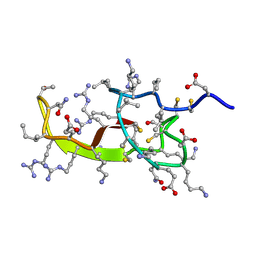 | |
4WCJ
 
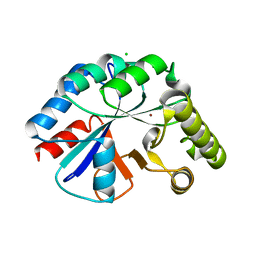 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
1D83
 
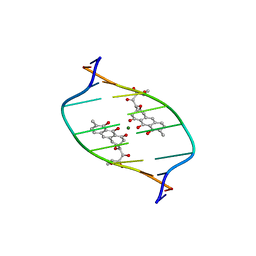 | | STRUCTURE REFINEMENT OF THE CHROMOMYCIN DIMER/DNA OLIGOMER COMPLEX IN SOLUTION | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Gao, X, Mirau, P, Patel, D.J. | | Deposit date: | 1992-07-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the chromomycin dimer-DNA oligomer complex in solution.
J.Mol.Biol., 223, 1992
|
|
7A10
 
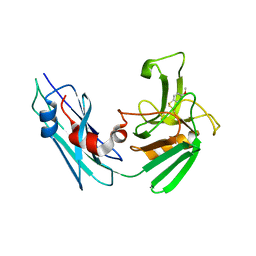 | | LppS with covalent adduct derived from 1g | | Descriptor: | 4-methoxycyclohexa-2,5-diene-1-thione, L,D-transpeptidase 2 | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A1E
 
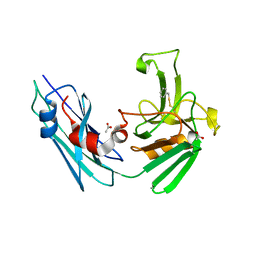 | | LppS with covalent adduct derived from 1c | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, phenylmethanethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A11
 
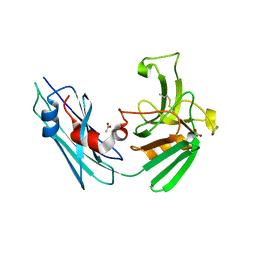 | | LppS with covalent adduct derived from 1E | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, propane-1-thiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A0Z
 
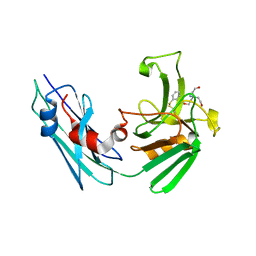 | | LppS with covalent adduct derived from 1b | | Descriptor: | L,D-transpeptidase 2, TRIS(HYDROXYETHYL)AMINOMETHANE, benzenethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A1C
 
 | |
3K23
 
 | | Glucocorticoid Receptor with Bound D-prolinamide 11 | | Descriptor: | 1-{[3-(4-{[(2R)-4-(5-fluoro-2-methoxyphenyl)-2-hydroxy-4-methyl-2-(trifluoromethyl)pentyl]amino}-6-methyl-1H-indazol-1-yl)phenyl]carbonyl}-D-prolinamide, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K0T
 
 | |
3K5X
 
 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
7S3W
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S3U
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S44
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S43
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
