4JGT
 
 | | Structure and kinetic analysis of H2S production by human Mercaptopyruvate Sulfurtransferase | | Descriptor: | 3-mercaptopyruvate sulfurtransferase, GLYCEROL, PYRUVIC ACID, ... | | Authors: | Koutmos, M, Yamada, K, Yadav, P.K, Chiku, T, Banerjee, R. | | Deposit date: | 2013-03-03 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Structure and Kinetic Analysis of H2S Production by Human Mercaptopyruvate Sulfurtransferase.
J.Biol.Chem., 288, 2013
|
|
4JQA
 
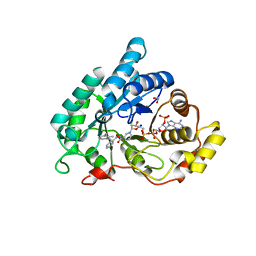 | | AKR1C2 complex with mefenamic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
4JKK
 
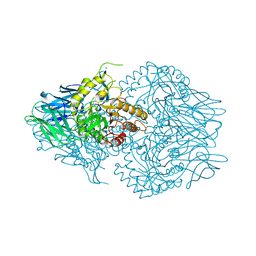 | |
4JR8
 
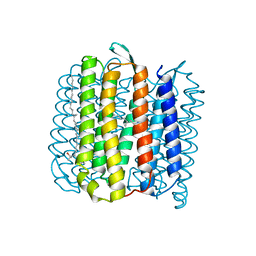 | |
4JSL
 
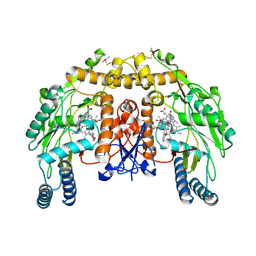 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6,6'-(heptane-1,7-diyl)bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-heptane-1,7-diylbis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In search of potent and selective inhibitors of neuronal nitric oxide synthase with more simple structures.
Bioorg.Med.Chem., 21, 2013
|
|
4JTQ
 
 | |
4J7C
 
 | |
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
4JC1
 
 | |
4JIE
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL, ... | | Authors: | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IEM
 
 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | Authors: | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3936 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
4IG0
 
 | | HIV-1 reverse transcriptase with bound fragment at the 507 site | | Descriptor: | 2-({[2-(3,4-dihydroquinolin-1(2H)-yl)-2-oxoethyl](methyl)amino}methyl)quinazolin-4(1H)-one, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4ILC
 
 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IMT
 
 | | Structure of rat neuronal nitric oxide synthase in complex with 6,6'-((4-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[4-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of selective inhibitors of neuronal nitric oxide synthase.
J.Med.Chem., 56, 2013
|
|
4IIL
 
 | | Crystal Structure of RfuA (TP0298) of T. pallidum Bound to Riboflavin | | Descriptor: | 1,2-ETHANEDIOL, Membrane lipoprotein TpN38(b), POTASSIUM ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes.
MBio, 4, 2013
|
|
4ILS
 
 | | Crystal structure of engineered protein. northeast structural genomics Consortium target or117 | | Descriptor: | Engineered protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of engineered protein. northeast structural genomics Consortium target or117
To be Published
|
|
4IMS
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), ACETATE ION, CHLORIDE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of selective inhibitors of neuronal nitric oxide synthase.
J.Med.Chem., 56, 2013
|
|
4IG3
 
 | | HIV-1 reverse transcriptase with bound fragment near Knuckles site | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IFY
 
 | | HIV-1 reverse transcriptase with bound fragment at the Knuckles site | | Descriptor: | 1-[4-(trifluoromethoxy)phenyl]methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IL1
 
 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
4IMW
 
 | | Structure of rat neuronal nitric oxide synthase in complex with 3,5-bis(2-(6-amino-4-methylpyridin-2-yl)ethyl)benzonitrile | | Descriptor: | 3,5-bis[2-(6-amino-4-methylpyridin-2-yl)ethyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of selective inhibitors of neuronal nitric oxide synthase.
J.Med.Chem., 56, 2013
|
|
4IN5
 
 | | (M)L214G mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Rhodobacter sphaeroides Photosynthetic Reaction Center Residue M214 in the Composition, Absorbance Properties, and Conformations of HA and BA Cofactors.
Biochemistry, 52, 2013
|
|
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4J7L
 
 | |
4JS5
 
 | |
