8RBO
 
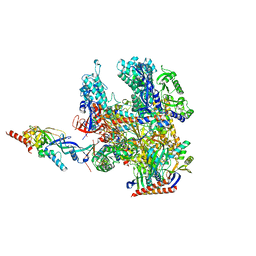 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
5C3E
 
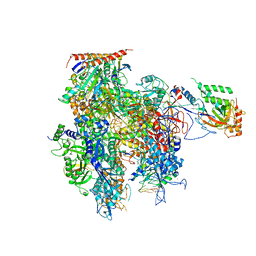 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
7XH9
 
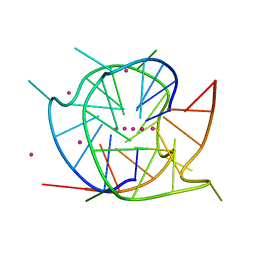 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*CP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION, STRONTIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XHD
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XIE
 
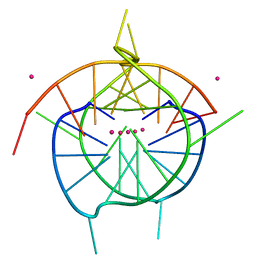 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*GP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7AZ5
 
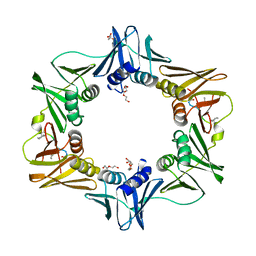 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
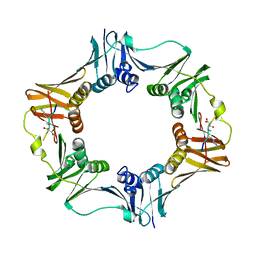 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
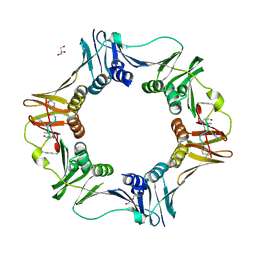 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
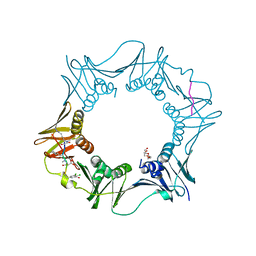 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
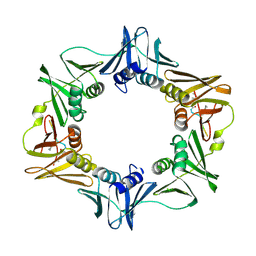 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, Peptide 22 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZF
 
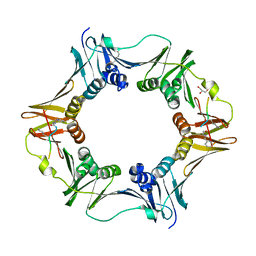 | | DNA polymerase sliding clamp from Escherichia coli with peptide 8 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZK
 
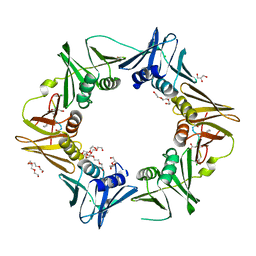 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
5OLA
 
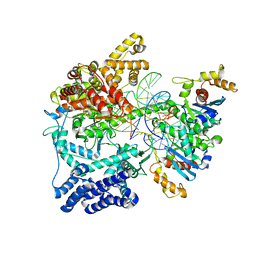 | | Structure of mitochondrial transcription elongation complex in complex with elongation factor TEFM | | Descriptor: | DNA (30-MER), DNA (5'-D(P*AP*TP*GP*GP*TP*GP*TP*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*GP*AP*AP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.904 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
6NSN
 
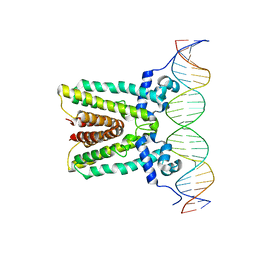 | |
7Q3Z
 
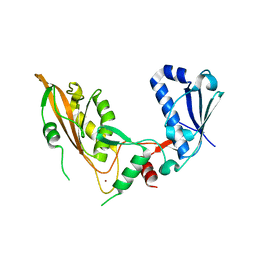 | | DNA/RNA binding protein | | Descriptor: | SODIUM ION, Schlafen family member 5, ZINC ION | | Authors: | Huber, E, Lammens, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
4LJZ
 
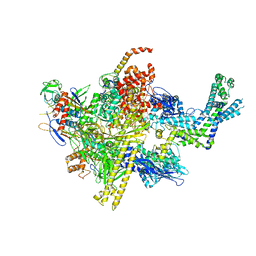 | | Crystal Structure Analysis of the E.coli holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5871 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5TBZ
 
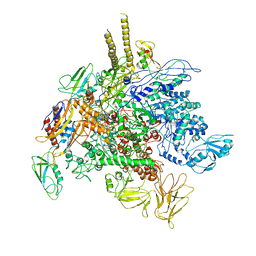 | | E. Coli RNA Polymerase complexed with NusG | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Steitz, T.A. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural insights into NusG regulating transcription elongation.
Nucleic Acids Res., 45, 2017
|
|
4BBR
 
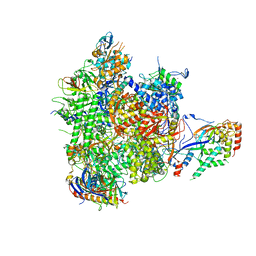 | | Structure of RNA polymerase II-TFIIB complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Sainsbury, S, Niesser, J, Cramer, P. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and Function of the Initially Transcribing RNA Polymerase II-TFIIB Complex
Nature, 493, 2013
|
|
6UPH
 
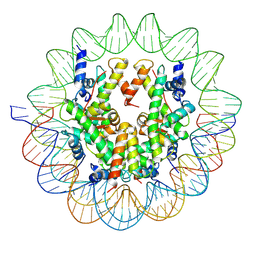 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | Descriptor: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | Authors: | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
8UPF
 
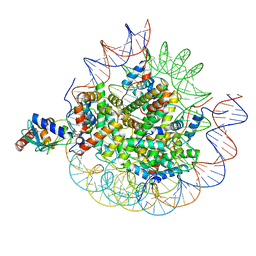 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
7YP9
 
 | |
