2MAJ
 
 | |
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2LX1
 
 | |
2M3S
 
 | | Calmodulin, i85l, f92e, h107i, l112r, a128t, m144r mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Moroz, Y.S, Wu, Y, Cheng, H, Roder, H, Korendovych, I.V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A single mutation in a regulatory protein produces evolvable allosterically regulated catalyst of nonnatural reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2M74
 
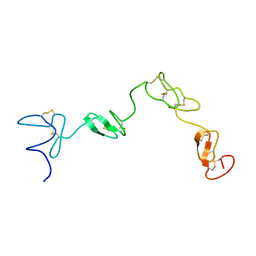 | | 1H, 13C and 15N assignments of the four N-terminal domains of human fibrillin-1 | | Descriptor: | Fibrillin-1 | | Authors: | Yadin, D.A, Robertson, I.B, Jensen, S.A, Handford, P.A, Redfield, C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-09-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Fibrillin-1 N-Terminal Domains Suggests that Heparan Sulfate Regulates the Early Stages of Microfibril Assembly.
Structure, 21, 2013
|
|
1WUZ
 
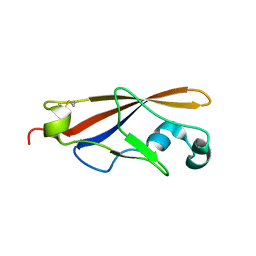 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
2MIT
 
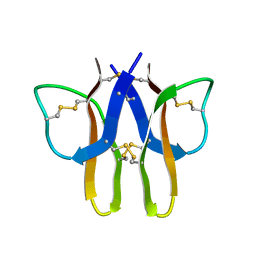 | |
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2M1Z
 
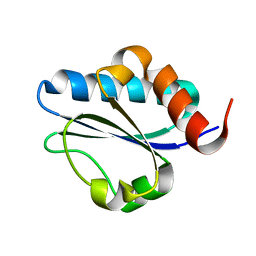 | |
2LXX
 
 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 2, isoform c | | Authors: | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
2LUF
 
 | |
2M1I
 
 | |
2M7V
 
 | | Green Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, PHYCOVIOLOBILIN | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2M7U
 
 | | Blue Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2MBF
 
 | |
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2MF7
 
 | | Solution structure of the ims domain of the mitochondrial import protein TIM21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bajaj, R, Jaremko, L, Jaremko, M, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space.
Structure, 22, 2014
|
|
2MJL
 
 | | Solution structure of peptidyl-tRNA hyrolase from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kabra, A, Shahid, S, Yadav, R, Pulavarti, S, Shukla, V.K, Arora, A. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptidyl-tRNA hydrolase from Vibrio cholerae
To be Published
|
|
2LVQ
 
 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2M8G
 
 | |
2N3T
 
 | |
2MTG
 
 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MUL
 
 | | Solution Structure of the UBM1 domain of human HUWE1/ARF-BP1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Farhadi, S, Khatun, R, Lemak, A, Kaustov, L, Ramabadran, R, Hunter, H, Sheng, Y. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin Binding Motif of human Arf-bp1
To be Published
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MPT
 
 | | WW3 domain of Nedd4L in complex with its HECT domain PY motif | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Escobedo, A, Macias, M.J, Gomes, T, Aragon, E, Martin-Malpartida, P, Ruiz, L. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Activation and Degradation Mechanisms of the E3 Ubiquitin Ligase Nedd4L.
Structure, 22, 2014
|
|
