3CZ0
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HOK
 
 | |
1TU0
 
 | | Aspartate Transcarbamoylase Catalytic Chain Mutant E50A Complex with Phosphonoacetamide | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, PHOSPHONOACETAMIDE, ... | | Authors: | Stieglitz, K, Stec, B, Baker, D.P, Kantrowitz, E.R. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
J.Mol.Biol., 341, 2004
|
|
1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TTH
 
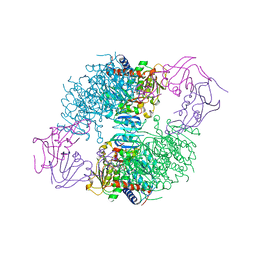 | | Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50Ala Complexed with N-(Phosphonacetyl-L-Aspartate) (PALA) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Stieglitz, K, Stec, B, Baker, D.P, Kantrowitz, E.R. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
J.Mol.Biol., 341, 2004
|
|
2HNY
 
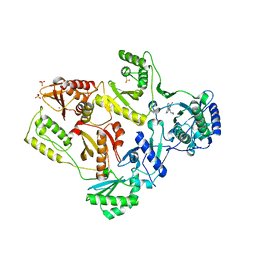 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HOM
 
 | |
2HOL
 
 | |
2HOO
 
 | |
2HOJ
 
 | |
2HOP
 
 | |
1YWL
 
 | |
4I7F
 
 | |
4ICL
 
 | | HIV-1 reverse transcriptase with bound fragment at the incoming dNTP binding site | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IFY
 
 | | HIV-1 reverse transcriptase with bound fragment at the Knuckles site | | Descriptor: | 1-[4-(trifluoromethoxy)phenyl]methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4EZ3
 
 | | CDK2 in complex with NSC 134199 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(6-hydroxy-2-oxo-1,2-dihydropyridin-3-yl)diazenyl]benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Martin, M, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
4IFV
 
 | | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, Exoribonuclease H, ... | | Authors: | Bauman, J.D, Patel, D, Fromer, M, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4ID5
 
 | | HIV-1 reverse transcriptase with bound fragment at the RNase H primer grip site | | Descriptor: | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
1RYL
 
 | | The Crystal Structure of a Protein of Unknown Function YfbM from Escherichia coli | | Descriptor: | Hypothetical protein yfbM | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a hypothetical protein yfbM from E. coli
To be Published
|
|
2FEO
 
 | | Mutant R188M of The Cytidine Monophosphate Kinase from E. coli complexed with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase, SULFATE ION | | Authors: | Ofiteru, A, Bucurenci, N, Alexov, E, Bertrand, T, Briozzo, P, Munier-Lehmann, H, Tourneux, L, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase.
Febs J., 274, 2007
|
|
3QJH
 
 | | The crystal structure of the 5c.c7 TCR | | Descriptor: | 5c.c7 alpha chain, 5c.c7 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QJF
 
 | | Crystal structure of the 2B4 TCR | | Descriptor: | 2B4 alpha chain, 2B4 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
4IDK
 
 | | HIV-1 reverse transcriptase with bound fragment at the 428 site | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-12 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
3ZG4
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
