1KMC
 
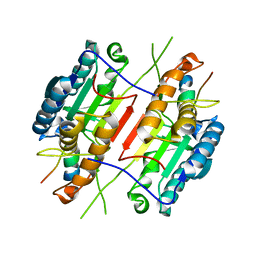 | |
1KMD
 
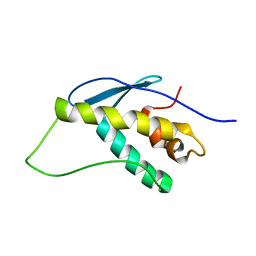 | | SOLUTION STRUCTURE OF THE VAM7P PX DOMAIN | | Descriptor: | Vacuolar morphogenesis protein VAM7 | | Authors: | Lu, J, Garcia, J, Dulubova, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Vam7p PX domain.
Biochemistry, 41, 2002
|
|
1KME
 
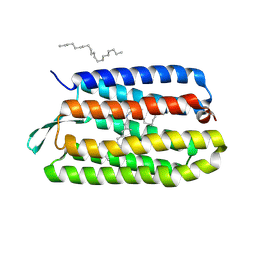 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN CRYSTALLIZED FROM BICELLES | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, RETINAL, ... | | Authors: | Faham, S, Bowie, J.U. | | Deposit date: | 2001-12-14 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure.
J.Mol.Biol., 316, 2002
|
|
1KMF
 
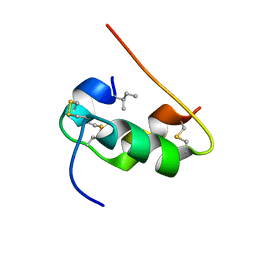 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
1KMH
 
 | |
1KMI
 
 | | CRYSTAL STRUCTURE OF AN E.COLI CHEMOTAXIS PROTEIN, CHEZ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BICINE, Chemotaxis protein cheY, ... | | Authors: | Zhao, R, Collins, E.J, Bourret, R.B, Silversmith, R.E. | | Deposit date: | 2001-12-16 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.
Nat.Struct.Biol., 9, 2002
|
|
1KMJ
 
 | |
1KMK
 
 | |
1KMM
 
 | |
1KMN
 
 | |
1KMO
 
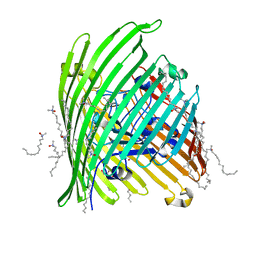 | | Crystal structure of the Outer Membrane Transporter FecA | | Descriptor: | HEPTANE-1,2,3-TRIOL, Iron(III) dicitrate transport protein fecA, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KMQ
 
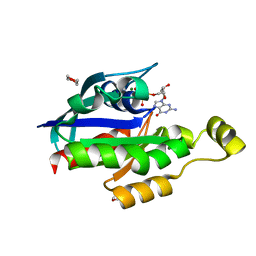 | | Crystal Structure of a Constitutively Activated RhoA Mutant (Q63L) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Longenecker, K, Read, P, Lin, S.-K, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a constitutively activated RhoA mutant (Q63L) at 1.55 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KMR
 
 | |
1KMS
 
 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9439), A LIPOPHILIC ANTIFOLATE | | Descriptor: | 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KMV
 
 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9662), A LIPOPHILIC ANTIFOLATE | | Descriptor: | (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, DIMETHYL SULFOXIDE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
1KMX
 
 | |
1KMY
 
 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 2,3-dihydroxybiphenyl under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KN0
 
 | | Crystal Structure of the human Rad52 protein | | Descriptor: | Rad52 | | Authors: | Kagawa, W, Kurumizaka, H, Ishitani, R, Fukai, S, Nureki, O, Shibata, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-18 | | Release date: | 2002-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form.
Mol.Cell, 10, 2002
|
|
1KN1
 
 | | Crystal structure of allophycocyanin | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Liang, D.C, Liu, J.Y, Jiang, T, Zhang, J.P, Chang, W.R. | | Deposit date: | 2001-12-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Allophycocyanin from red algae Porphyra yezoensis at 2.2 A resolution
J.BIOL.CHEM., 274, 1999
|
|
1KN2
 
 | | CATALYTIC ANTIBODY D2.3 COMPLEX | | Descriptor: | IG ANTIBODY D2.3 (HEAVY CHAIN), IG ANTIBODY D2.3 (LIGHT CHAIN), PARA-NITROPHENYL PHOSPHONOBUTANOYL L-ALANINE, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remarkable remote chiral recognition in a reaction mediated by a catalytic antibody.
J.Am.Chem.Soc., 124, 2002
|
|
1KN3
 
 | |
