8HY8
 
 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY9
 
 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.462 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
3MHS
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
8HZS
 
 | |
4CWM
 
 | | High-glycosylation crystal structure of the bifunctional endonuclease (AtBFN2) from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDONUCLEASE 2, ZINC ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4PSH
 
 | | Structure of holo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3, ARGININE | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
3ML0
 
 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5YHA
 
 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
4L15
 
 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
3MLT
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a UG1033 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MPS
 
 | | Peroxide Bound Oxidized Rubrerythrin from Pyrococcus furiosus | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, MU-OXO-DIIRON, ... | | Authors: | Dillard, B.D, Adams, M.W.W, Lanzilotta, W.N. | | Deposit date: | 2010-04-27 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | Descriptor: | Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
4PJL
 
 | | Myosin VI motor domain A458E mutant in the Pi release state, space group P212121 - | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
5Y8E
 
 | |
1FGB
 
 | | TOXIN | | Descriptor: | CHOLERA TOXIN B SUBUNIT PENTAMER | | Authors: | Zhang, R.-G, Westbrook, E. | | Deposit date: | 1996-02-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.4 A crystal structure of cholera toxin B subunit pentamer: choleragenoid.
J.Mol.Biol., 251, 1995
|
|
3EFV
 
 | | Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-04 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
7TCI
 
 | | Structure of Xenopus KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, CALCIUM ION, Calmodulin-1, ... | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
4PRS
 
 | | Structure of apo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3 | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
4PV1
 
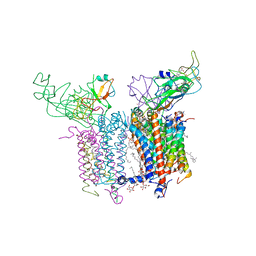 | | Cytochrome B6F structure from M. laminosus with the quinone analog inhibitor stigmatellin | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Hasan, S.S, Yamashita, E, Cramer, W.A. | | Deposit date: | 2014-03-14 | | Release date: | 2014-08-20 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Traffic within the cytochrome b6f lipoprotein complex: gating of the quinone portal.
Biophys.J., 107, 2014
|
|
3M5O
 
 | |
3M08
 
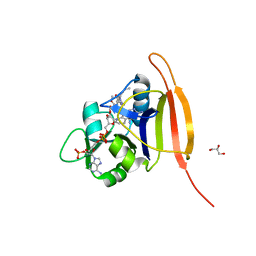 | | Wild Type Dihydrofolate Reductase from Staphylococcus aureus with inhibitor RAB1 | | Descriptor: | 5-(3,4-dimethoxy-5-{(1E)-3-oxo-3-[(1S)-1-propylphthalazin-2(1H)-yl]prop-1-en-1-yl}benzyl)pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.014 Å) | | Cite: | Inhibition of Antibiotic-Resistant Staphylococcus aureus by the Broad-Spectrum Dihydrofolate Reductase Inhibitor RAB1.
Antimicrob.Agents Chemother., 54, 2010
|
|
5BWM
 
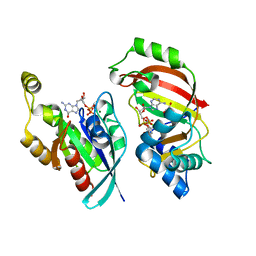 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-06-08 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
7TCP
 
 | | Structure of Xenopus KCNQ1-CaM | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
3M1C
 
 | |
2J8Y
 
 | | Structure of PBP-A acyl-enzyme complex with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-10-31 | | Release date: | 2007-11-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
