3U2O
 
 | | Dihydroorotate Dehydrogenase (DHODH) crystal structure in complex with small molecule inhibitor | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Lozoya, E, Segarra, V, Erra, M, Wenzkowski, C, Jestel, A, Krapp, S, Blaesse, M. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biaryl analogues of teriflunomide as potent DHODH inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3U6A
 
 | | Rational Design and Synthesis of Aminopiperazinones as Beta Secretase (BACE) Inhibitors | | Descriptor: | Beta-secretase 1, N-{3-[(2R)-6-amino-2,4-dimethyl-3-oxo-2,3,4,5-tetrahydropyrazin-2-yl]phenyl}-5-chloropyridine-2-carboxamide, SULFATE ION | | Authors: | Spurlino, J.C, Alexander, R.S. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-09 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Rational design and synthesis of aminopiperazinones as beta-secretase (BACE) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2R4F
 
 | | Substituted Pyrazoles as Hepatselective HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[1-(4-fluorophenyl)-4-(1-methylethyl)-3-{methyl[(1R)-1-phenylethyl]carbamoyl}-1H-pyrazol-5-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors: discovery of (3R,5R)-7-[2-(4-fluoro-phenyl)-4-isopropyl-5-(4-methyl-benzylcarbamoyl)-2H-pyrazol-3-yl]-3,5-dihydroxyheptanoic acid (PF-3052334) as a candidate for the treatment of hypercholesterolemia.
J.Med.Chem., 51, 2008
|
|
3PJC
 
 | | Crystal structure of JAK3 complexed with a potent ATP site inhibitor showing high selectivity within the Janus kinase family | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-oxopiperidin-1-yl)-5-(trifluoromethyl)pyrimidin-4-yl]-1H-pyrrole-2,5-dione, Tyrosine-protein kinase JAK3 | | Authors: | Tavares, G.A, Thoma, G, Zerwes, H.-G, Kroemer, M. | | Deposit date: | 2010-11-09 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Potent Janus Kinase 3 Inhibitor with High Selectivity within the Janus Kinase Family.
J.Med.Chem., 54, 2011
|
|
4G20
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3LDL
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
4GL5
 
 | |
4G21
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3LDO
 
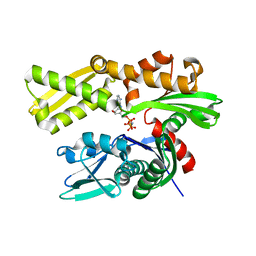 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMPPNP | | Descriptor: | 78 kDa glucose-regulated protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3EKN
 
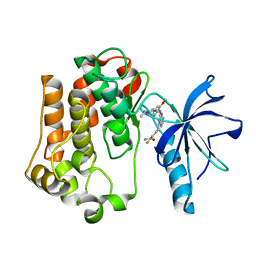 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[4-(1-methylethyl)piperazin-1-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4G1Y
 
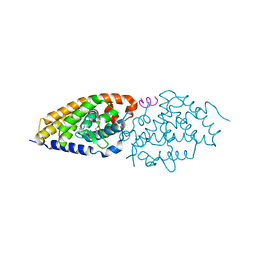 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (4E,6Z)-7-(3-{[3,4-bis(hydroxymethyl)benzyl]oxy}phenyl)-3-ethylnona-4,6-dien-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3EKK
 
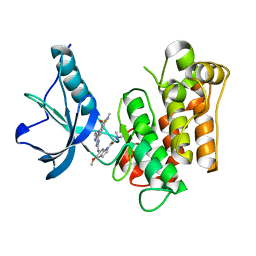 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-[(2-{[1-(N,N-dimethylglycyl)-5-methoxy-1H-indol-6-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-6-fluoro-N-methylbenzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Sabbatini, P, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidines: Potent inhibitors of the IGF-1R receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EMG
 
 | |
4ZRO
 
 | | 2.1 A X-Ray Structure of FIPV-3CLpro bound to covalent inhibitor | | Descriptor: | 3C-like proteinase, Bounded inhibitor of N-(tert-butoxycarbonyl)-L-seryl-L-valyl-N-{(2S)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-leucinamide, DIMETHYL SULFOXIDE | | Authors: | St John, S.E, Mesecar, A.D. | | Deposit date: | 2015-05-12 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.0566 Å) | | Cite: | X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: Structural implications for drug design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3EQP
 
 | | Crystal Structure of Ack1 with compound T95 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N-(2,6-dimethylphenyl)-4-(2-ethoxyphenoxy)-2-({4-[4-(2-hydroxyethyl)piperazin-1-yl]phenyl}amino)pyrimidine-5-carboxamide | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZM4
 
 | |
5ADQ
 
 | | Crystal structure of human tankyrase 2 in complex with JW55 | | Descriptor: | BICARBONATE ION, GLYCEROL, N-(4-(((4-(4-methoxyphenyl)oxan-4- yl)methyl)carbamoyl)phenyl)furan-2-carboxamide, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5ADS
 
 | |
5ADR
 
 | | Crystal structure of human tankyrase 2 in complex with OD38 | | Descriptor: | BICARBONATE ION, N-(4-(((4-(4-methoxyphenyl)oxan-4- yl)methyl)carbamoyl)phenyl)-5-methylfuran-2-carboxamide, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3NUP
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(1-methylpiperidin-4-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
3NUX
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[5-chloro-3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(5-piperazin-1-ylpyridin-2-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
5ADT
 
 | | Crystal structure of human tankyrase 2 in complex with OD73 | | Descriptor: | BICARBONATE ION, N-[3-chloranyl-4-[[4-(4-methoxyphenyl)oxan-4-yl]methylcarbamoyl]phenyl]-2-methyl-1,3-oxazole-5-carboxamide, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4HMN
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (4-(4-Chlorophenyl)piperazin-1-yl)(morpholino)methanone (24) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Turnbull, A.P, Flanagan, J.U, Atwell, G.J, Heinrich, D.M, Jamieson, S.M.F, Brooke, D.G, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Morpholylureas are a new class of potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase (AKR1C3).
Bioorg.Med.Chem., 22, 2014
|
|
3RVI
 
 | |
3O2A
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
