4YTH
 
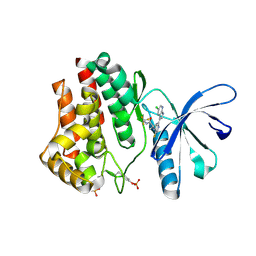 | |
8ATH
 
 | | CRYSTAL STRUCTURE OF LAMP1 IN COMPLEX WITH FAB-B. | | Descriptor: | Fab B Heavy Chain, Fab B Light Chain, Lysosome-associated membrane glycoprotein 1 | | Authors: | Mathieu, M, Dupuy, A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.366 Å) | | Cite: | Deciphering cross-species reactivity of LAMP-1 antibodies using deep mutational epitope mapping and AlphaFold.
Mabs, 15, 2023
|
|
6ZI3
 
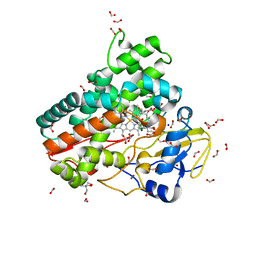 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6YYN
 
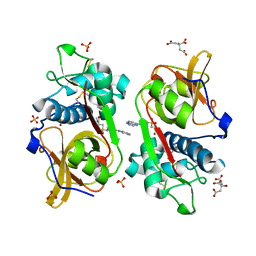 | | Structure of Cathepsin S in complex with Compound 14 | | Descriptor: | CITRATE ANION, Cathepsin S, SULFATE ION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYO
 
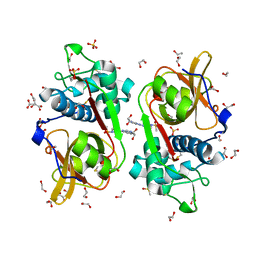 | | Structure of Cathepsin S in complex with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylsulfonylpiperazin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6U50
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
3DRT
 
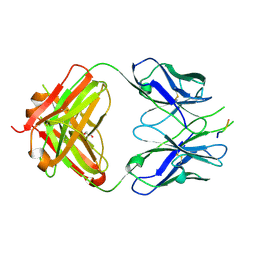 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
3DJ6
 
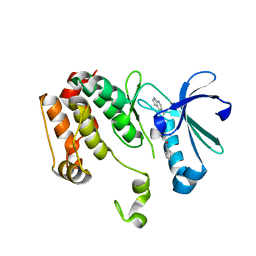 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 823. | | Descriptor: | 4-methoxy-N-phenyl-3-({3-[(1H-pyrrolo[2,3-b]pyridin-5-ylmethyl)sulfanyl]propanoyl}amino)benzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
8G8Y
 
 | |
5LAW
 
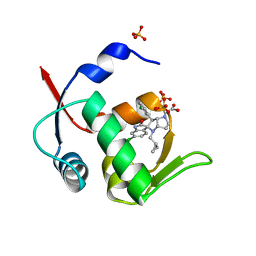 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | Descriptor: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAZ
 
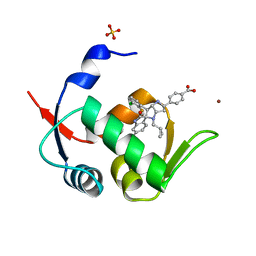 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | Descriptor: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
3EGS
 
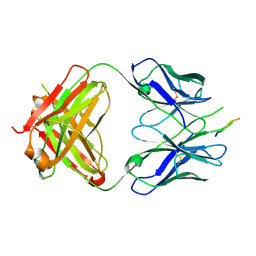 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 soaked in ammonium sulfate | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, gp41 scrFP-MPER construct | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
8EE0
 
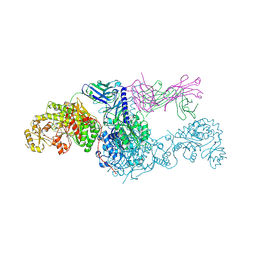 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment 1B2 | | Descriptor: | 1B2 antibody heavy chain, 1B2 antibody light chain, 6-deoxyerythronolide B synthase | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
8CS9
 
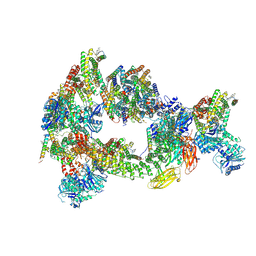 | | Composite reconstruction of Class 1 of the erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6OL6
 
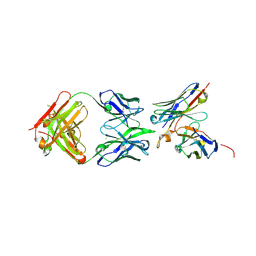 | | Structure of iglb12 scFv in complex with anti-idiotype ib2 Fab | | Descriptor: | CHLORIDE ION, ib2 Heavy Chain, ib2 Light Chain, ... | | Authors: | Weidle, C, Pancera, M, Gewe, M. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes.
J.Exp.Med., 216, 2019
|
|
7Q0Q
 
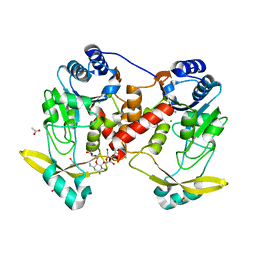 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 3N methyl nemycin B, ACETATE ION, Aminoglycoside N(3)-acetyltransferase III, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-16 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The 3-N-alkylation of the neomycin B outmaneuvers the aminoglycoside resistant enzyme acetyltransferase(3)IIIa via an unexpected mechanism
To Be Published
|
|
7QRC
 
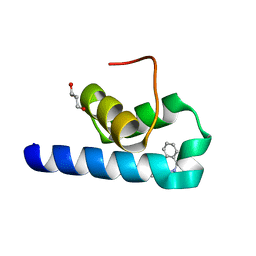 | | X-ray structure of Trypanosoma cruzi PEX14 in complex with a PEX5-PEX14 PPI inhibitor | | Descriptor: | GLYCEROL, Peroxin-14, ~{N}-(5-ethyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)-2-(4-fluorophenyl)ethanamide | | Authors: | Napolitano, V, Popowicz, G.M, Dawidowski, M, Dubin, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based design, synthesis and evaluation of a novel family of PEX5-PEX14 interaction inhibitors against Trypanosoma.
Eur.J.Med.Chem., 243, 2022
|
|
8OWA
 
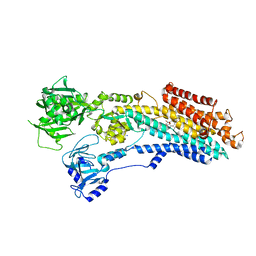 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-4 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8OWL
 
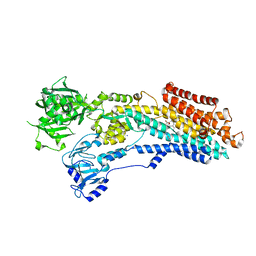 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-6 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8PDK
 
 | | X-ray structure of the Thermus thermophilus PilF-GSPIIB domain in the c-di-GMP bound state | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, SULFATE ION, ... | | Authors: | Neissner, K, Woehnert, J. | | Deposit date: | 2023-06-12 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for high-affinity c-di-GMP binding to the GSPII-B domain of the traffic ATPase PilF from Thermus thermophilus.
J.Biol.Chem., 301, 2024
|
|
6LZB
 
 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with AN-P2-5H-06 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]-~{N}-oxidanyl-pyrrolo[2,3-b]pyridine-5-carboxamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
6LZC
 
 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with KV-P2-4H-05 | | Descriptor: | COBALT (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
6XV7
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N},3-dimethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XKU
 
 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKZ
 
 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
