5WG8
 
 | | Structure of PP5C with LB-100; 7-oxabicyclo[2.2.1]heptane-2,3-dicarbonyl moiety modeled in the density | | Descriptor: | (1S,2R,3S,4R)-3-(4-methylpiperazine-1-carbonyl)-7-oxabicyclo[2.2.1]heptane-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | D'Arcy, B.M, Swingle, M.R, Honkanen, R.E, Prakash, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Antitumor Drug LB-100 Is a Catalytic Inhibitor of Protein Phosphatase 2A (PPP2CA) and 5 (PPP5C) Coordinating with the Active-Site Catalytic Metals in PPP5C.
Mol. Cancer Ther., 18, 2019
|
|
5W0W
 
 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
3P71
 
 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
3LL8
 
 | |
4V0W
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-669) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
7CUN
 
 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
7DOG
 
 | |
4WWL
 
 | | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form) | | Descriptor: | CARBONATE ION, GLYCEROL, Protein UshA, ... | | Authors: | Krug, U, Paithankar, K.S, Schultz-Heienbrok, R, Strater, N. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form)
To Be Published
|
|
4DT2
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC27209 | | Descriptor: | (2,2-dimethyl-2,3-dihydro-1-benzofuran-7-yl)methanol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
5UI1
 
 | | Crystal Structure of Human Protein Phosphatase 5C (PP5C) in complex with a triazole inhibitor | | Descriptor: | 5-phenyl-1H-1,2,3-triazole-4-carboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Banerjee, S, Honkanen, R.E. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure Human PP5C in Complex with an Inhibitor
To Be Published
|
|
4XPN
 
 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
4DHL
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment MO07123 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Clayton, D.J, Hussein, W.M, Schenk, G, McGeary, R, Guddat, L.W. | | Deposit date: | 2012-01-29 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
5SVE
 
 | | Structure of Calcineurin in complex with NFATc1 LxVP peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Sheftic, S.R, Page, R, Peti, W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Investigating the human Calcineurin Interaction Network using the pi LxVP SLiM.
Sci Rep, 6, 2016
|
|
1FJM
 
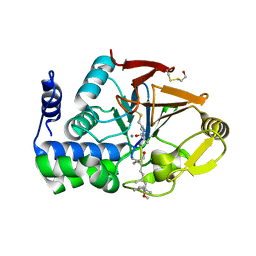 | | Protein serine/threonine phosphatase-1 (alpha isoform, type 1) complexed with microcystin-LR toxin | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, PROTEIN SERINE/THREONINE PHOSPHATASE-1 (ALPHA ISOFORM, ... | | Authors: | Goldberg, J, Nairn, A.C, Kuriyan, J. | | Deposit date: | 1995-12-17 | | Release date: | 1996-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1.
Nature, 376, 1995
|
|
3ZTV
 
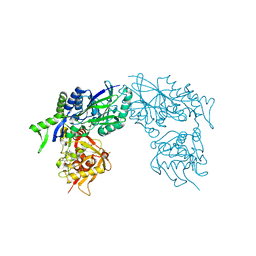 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | ADENOSINE, GLYCEROL, NAD NUCLEOTIDASE, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
3ZU0
 
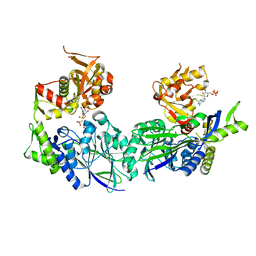 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | NAD NUCLEOTIDASE, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
6HMS
 
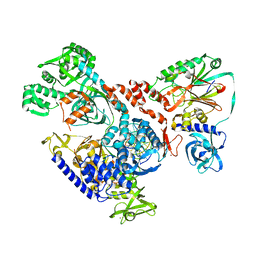 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
6HWR
 
 | | Red kidney bean purple acid phosphatase in complex with adenosine divanadate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Gahan, L.R, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2018-10-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Binding Mode of an ADP Analogue to a Metallohydrolase Mimics the Likely Transition State.
Chembiochem, 20, 2019
|
|
4YKE
 
 | |
6HXW
 
 | | structure of human CD73 in complex with antibody IPH53 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, IPH53 heavy chain, ... | | Authors: | Roussel, A, Amigues, B. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies.
Cell Rep, 27, 2019
|
|
4DSY
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
7BBJ
 
 | |
5UKI
 
 | |
3ZK4
 
 | |
3V4Y
 
 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
