7QHA
 
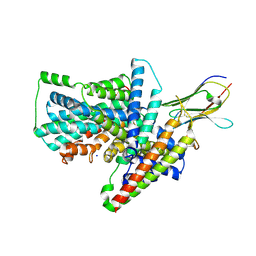 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | | Descriptor: | Megabody c7HopQ, Putative TRAP-type C4-dicarboxylate transport system, large permease component, ... | | Authors: | North, R.A, Davies, J.S, Morado, D, Dobson, R.C.J. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
5B4L
 
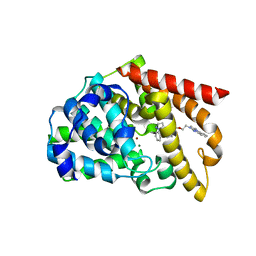 | | Crystal structure of the catalytic domain of human PDE10A complexed with 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
5B4K
 
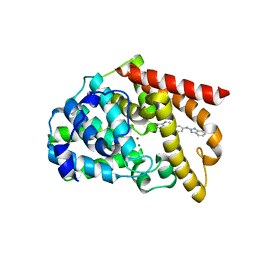 | | Crystal structure of the catalytic domain of human PDE10A complexed with N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine | | Descriptor: | MAGNESIUM ION, N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
4QTS
 
 | |
4XY2
 
 | | Crystal structure of PDE10A in complex with ASP9436 | | Descriptor: | 1-methyl-5-(1-methyl-3-{[4-(1-methyl-1H-benzimidazol-4-yl)phenoxy]methyl}-1H-pyrazol-4-yl)pyridin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2015-02-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Addressing phototoxicity observed in a novel series of biaryl derivatives: Discovery of potent, selective and orally active phosphodiesterase 10A inhibitor ASP9436
Bioorg.Med.Chem., 23, 2015
|
|
4QU4
 
 | |
7RZY
 
 | | CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Tn6677 Vibrio cholerae transposon TnsC (VchTnsC) | | Authors: | Hoffmann, F.T, Kim, M, Beh, L.Y, Wang, J, Vo, P.L.H, Gelsinger, D.R, Acree, C, Mohabir, J.T, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2021-08-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
1PHD
 
 | |
1PHG
 
 | |
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
5C2H
 
 | | PDE10 complexed with 6-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]-5-methyl-2-[3-(2-quinolyl)propoxy]pyrimidin-4-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 6-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-5-methyl-2-[3-(quinolin-2-yl)propoxy]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of a Series of Pyrimidine-Based Phosphodiesterase 10A (PDE10A) Inhibitors through Fragment Screening, Structure-Based Design, and Parallel Synthesis.
J.Med.Chem., 58, 2015
|
|
1N7S
 
 | | High Resolution Structure of a Truncated Neuronal SNARE Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, SNAP-25A, ... | | Authors: | Ernst, J.A, Brunger, A.T. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex
J.Biol.Chem., 278, 2003
|
|
1PHF
 
 | |
1PHE
 
 | |
4TPP
 
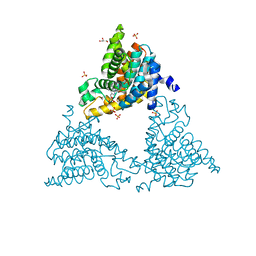 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
7TBG
 
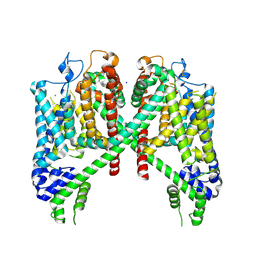 | | AtTPC1 D454N with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, SODIUM ION, Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-22 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDF
 
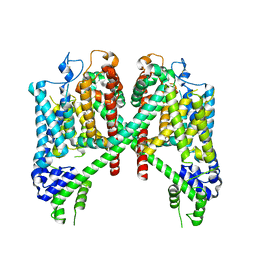 | | AtTPC1 D454N with 1 mM EDTA state I | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5DH5
 
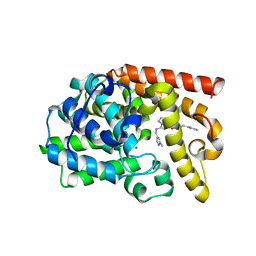 | | PDE10 complexed with N-[(1-methylpyrazol-4-yl)methyl]-5-[[(1S,2S)-2-(2-pyridyl)cyclopropyl]methoxy]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | MAGNESIUM ION, N-[(1-methyl-1H-pyrazol-4-yl)methyl]-5-{[(1S,2S)-2-(pyridin-2-yl)cyclopropyl]methoxy}pyrazolo[1,5-a]pyrimidin-7-amine, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1N2O
 
 | |
4TPM
 
 | |
6NUE
 
 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
8C38
 
 | |
3FAS
 
 | | X-ray structure of iGluR4 flip ligand-binding core (S1S2) in complex with (S)-glutamate at 1.40A resolution | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 4, ... | | Authors: | Kasper, C, Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2008-11-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of agonist recognition by the ligand-binding core of the ionotropic glutamate receptor 4
Febs Lett., 582, 2008
|
|
1N2I
 
 | | Crystal Structure of Pantothenate Synthetase from M. tuberculosis in complex with a reaction intermediate, pantoyl adenylate, different occupancies of pantoyl adenylate | | Descriptor: | ETHANOL, GLYCEROL, PANTOYL ADENYLATE, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-22 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a pantothenate
synthetase from M. tuberculosis and its
complexes with substrates and a
reaction intermediate
Protein Sci., 12, 2003
|
|
