2OFD
 
 | |
4I8D
 
 | | Crystal Structure of Beta-D-glucoside glucohydrolase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucoside glucohydrolase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Helmich, K.E, Banerjee, G, Bianchetti, C.M, Gudmundsson, M, Sandgren, M, Walton, J.D, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 beta-Glucosidase, Cel3A from Hypocrea jecorina.
J.Biol.Chem., 289, 2014
|
|
2EJN
 
 | | Structural characterization of the tetrameric form of the major cat allergen fel D 1 | | Descriptor: | CALCIUM ION, Major allergen I polypeptide chain 1, chain 2 | | Authors: | Kaiser, L, Velickovic, T.C, Badia-Martinez, D, Adedoyin, J, Thunberg, S, Hallen, D, Berndt, K, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the tetrameric form of the major cat allergen Fel d 1
J.Mol.Biol., 370, 2007
|
|
1EX1
 
 | | BETA-D-GLUCAN EXOHYDROLASE FROM BARLEY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-D-GLUCAN EXOHYDROLASE ISOENZYME EXO1), ... | | Authors: | Varghese, J.N, Hrmova, M, Fincher, G.B. | | Deposit date: | 1998-11-10 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a barley beta-D-glucan exohydrolase, a family 3 glycosyl hydrolase.
Structure Fold.Des., 7, 1999
|
|
1GPE
 
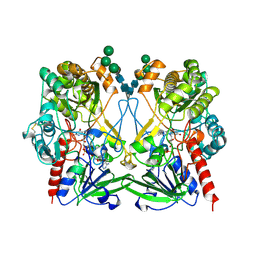 | | GLUCOSE OXIDASE FROM PENICILLIUM AMAGASAKIENSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hendle, J, Kalisz, H.M, Hecht, H.J. | | Deposit date: | 1999-03-24 | | Release date: | 1999-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CF3
 
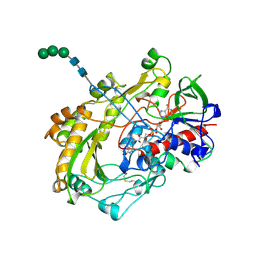 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2EXK
 
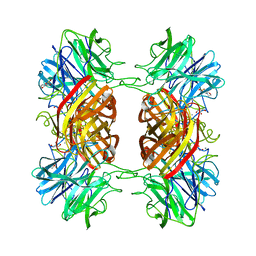 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
3U7B
 
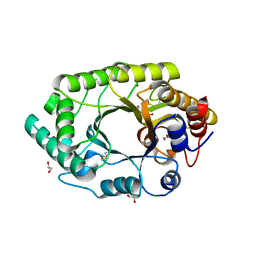 | | A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of a GH10 xylanase from Fusarium oxysporum reveals the presence of an extended loop on top of the catalytic cleft.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2EFU
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFX
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2HI8
 
 | | human formylglycine generating enzyme, C336S mutant, bromide co-crystallization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Rudolph, M.G, Roeser, D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HIB
 
 | | human formylglycine generating enzyme, C336S mutant, iodide co-crystallization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Rudolph, M.G, Roeser, D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2EXJ
 
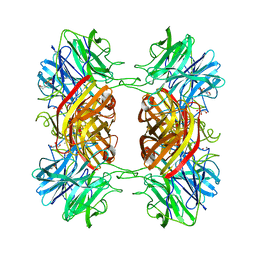 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2GYI
 
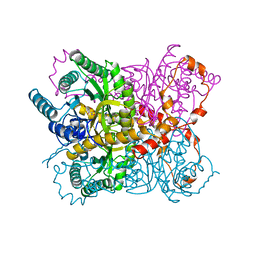 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
2ESC
 
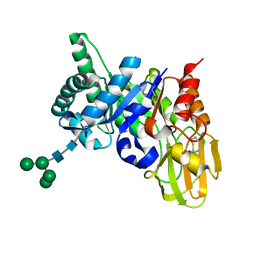 | | Crystal structure of a 40 KDa protective signalling protein from Bovine (SPC-40) at 2.1 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastav, D.B, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bovine secretory signalling glycoprotein (SPC-40) at 2.1 Angstrom resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7LSP
 
 | |
4LFL
 
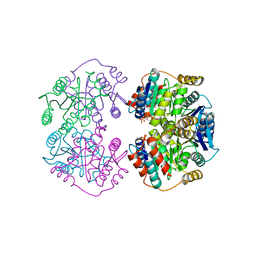 | |
1KQ0
 
 | | Human methionine aminopeptidase type II in complex with D-methionine | | Descriptor: | D-METHIONINE, Methionine aminopeptidase 2, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Nonato, M.C, Widom, J, Clardy, J. | | Deposit date: | 2002-01-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human methionine aminopeptidase type 2 in complex with L- and D-methionine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3CTL
 
 | | Crystal structure of D-Allulose 6-Phosphate 3-Epimerase from Escherichia coli K12 complexed with D-glucitol 6-phosphate and magnesium | | Descriptor: | D-SORBITOL-6-PHOSPHATE, D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
7SB8
 
 | | d(GA(CGA)5) parallel-stranded homo-duplex | | Descriptor: | COBALT HEXAMMINE(III), GA(CGA)5, SODIUM ION, ... | | Authors: | Luteran, E.M, Paukstelis, P.J. | | Deposit date: | 2021-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The parallel-stranded d(CGA) duplex is a highly predictable structural motif with two conformationally distinct strands.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2I57
 
 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
4UQC
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum at 1.30 A resolution | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
