3FDS
 
 | |
3VD0
 
 | | structure of p73 DNA binding domain tetramer modulates p73 transactivation | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*G)-3'), Tumor protein p73, ZINC ION | | Authors: | Ethayathulla, A.S, Tse, P.W, Nguyen, S, Viadiu, H. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4QTR
 
 | | Computational design of co-assembling protein-DNA nanowires | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*TP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*AP*TP*TP*TP*AP*AP*TP*TP*TP*CP*C)-3'), dualENH | | Authors: | Mou, Y, Mayo, S.L. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of co-assembling protein-DNA nanowires.
Nature, 525, 2015
|
|
3DFX
 
 | | Opposite GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
1A0A
 
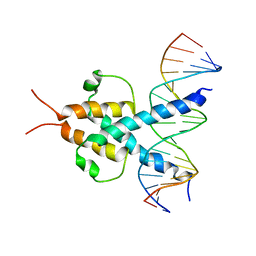 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
6O3T
 
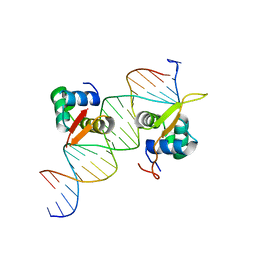 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
3M9E
 
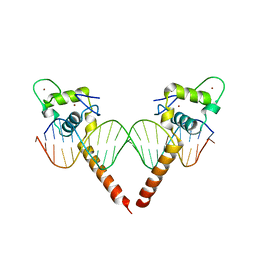 | | Thyroid hormone beta DNA binding domain homodimer with inverted palindrome TRE | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*AP*CP*CP*TP*CP*AP*GP*CP*TP*GP*AP*GP*GP*TP*CP*AP*AP*T)-3'), Thyroid hormone receptor beta, ZINC ION | | Authors: | Chen, Y. | | Deposit date: | 2010-03-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structure of a thyroid hormone receptor DNA-binding domain homodimer bound to an inverted palindrome DNA response element.
Mol.Endocrinol., 24, 2010
|
|
9DWI
 
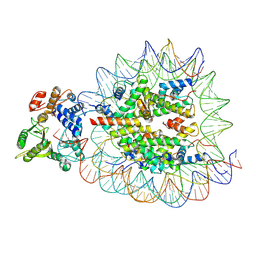 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 3, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 4, composite)
To Be Published
|
|
9DWH
 
 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 2, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 2, composite)
To Be Published
|
|
3VW4
 
 | | Crystal structure of the DNA-binding domain of ColE2-P9 Rep in complex with the replication origin | | Descriptor: | DNA (5'-D(P*AP*AP*TP*GP*AP*GP*AP*CP*CP*AP*GP*AP*TP*AP*AP*GP*CP*CP*TP*TP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*AP*AP*GP*GP*CP*TP*TP*AP*TP*CP*TP*GP*GP*TP*CP*TP*CP*AP*TP*T)-3'), Rep, ... | | Authors: | Itou, H, Yagura, M, Itoh, T, Shirakihara, Y. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
J.Biol.Chem., 290, 2015
|
|
2HAP
 
 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
4L62
 
 | |
8JRB
 
 | |
8SY7
 
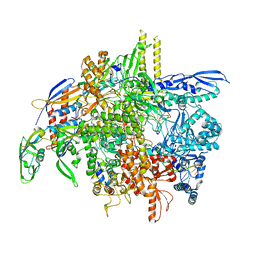 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
7PBK
 
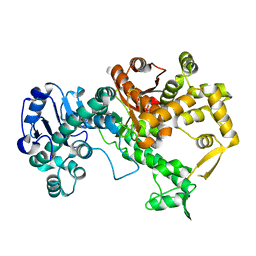 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
1BG1
 
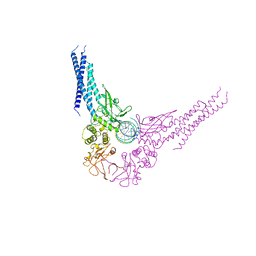 | | TRANSCRIPTION FACTOR STAT3B/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR STAT3B) | | Authors: | Becker, S, Groner, B, Muller, C.W. | | Deposit date: | 1998-06-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Three-dimensional structure of the Stat3beta homodimer bound to DNA.
Nature, 394, 1998
|
|
9DWK
 
 | |
9DWG
 
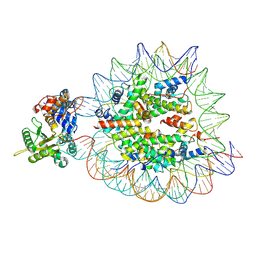 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite)
To Be Published
|
|
9DWM
 
 | |
1IF1
 
 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
6NJQ
 
 | | Structure of TBP-Hoogsteen containing DNA complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A, Stelling, A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Infrared Spectroscopic Observation of a G-C+Hoogsteen Base Pair in the DNA:TATA-Box Binding Protein Complex Under Solution Conditions.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5J0R
 
 | |
5J0X
 
 | |
5J0O
 
 | |
