4UWI
 
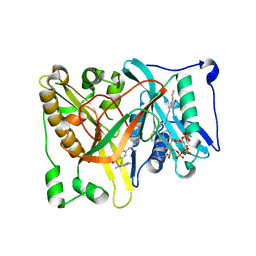 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-4-[3-(4-methylpiperazin-1-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4UVJ
 
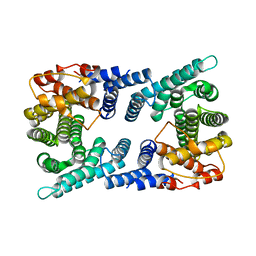 | |
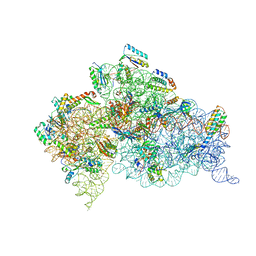
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2UUA
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
8H6B
 
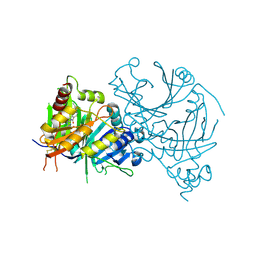 | | Crystal structure of AtHPPD complexed with YH20702 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-(1-methyl-1,2,3,4-tetrazol-5-yl)-1-pentyl-pyrrolo[2,3-b]pyridine-4-carboxamide | | Authors: | Yang, G.-F. | | Deposit date: | 2022-10-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Crystal structure of AtHPPD complexed with YH20702
To Be Published
|
|
2UTG
 
 | |
4V2Z
 
 | |
8HFD
 
 | | Crystal structure of allantoinase from E. coli BL21 | | Descriptor: | Allantoinase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Lin, E.S, Huang, H.Y, Yang, P.C, Liu, H.W, Huang, C.Y. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis.
Molecules, 28, 2023
|
|
4UTH
 
 | | XenA - oxidized - Y183F variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, SULFATE ION | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
2UUE
 
 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | 1-(3,5-DICHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOLE-3-CARBOXYLIC ACID, 4-METHYL-5-{(2E)-2-[(4-MORPHOLIN-4-YLPHENYL)IMINO]-2,5-DIHYDROPYRIMIDIN-4-YL}-1,3-THIAZOL-2-AMINE, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-03-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
8HKR
 
 | | Crystal Structure of Histone H3 Lysine 79 (H3K79) Methyltransferase Rv2067c from Mycobacterium tuberculosis | | Descriptor: | PHOSPHATE ION, Protein lysine methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dadireddy, V, Singh, P.R, Kalladi, S.M, Valakunja, N, Ramakumar, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Mycobacterium tuberculosis methyltransferase Rv2067c manipulates host epigenetic programming to promote its own survival.
Nat Commun, 14, 2023
|
|
4UVV
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methylisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
2UUR
 
 | | N-terminal NC4 domain of collagen IX | | Descriptor: | 1,2-ETHANEDIOL, COLLAGEN ALPHA-1(IX) CHAIN, SULFATE ION, ... | | Authors: | Leppanen, V.-M, Tossavainen, H, Permi, P, Lehtio, L, Ronnholm, G, Goldman, A, Kilpelainen, I, Pihlajamaa, T. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Terminal Nc4 Domain of Collagen Ix, a Zinc Binding Member of the Laminin-Neurexin-Sex Hormone Binding Globulin (Lns) Domain Family.
J.Biol.Chem., 282, 2007
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
8HO6
 
 | | ScRIPK WT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ScRIPK kinase protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
2UVJ
 
 | |
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8GYY
 
 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose, 120 seconds | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, L.Y. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
2UVS
 
 | | High Resolution Solid-state NMR structure of Kaliotoxin | | Descriptor: | POTASSIUM CHANNEL TOXIN ALPHA-KTX 3.1 | | Authors: | Korukottu, J, Lange, A, Vijayan, V, Schneider, R, Pongs, O, Becker, S, Baldus, M, Zweckstetter, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-05-27 | | Last modified: | 2020-01-15 | | Method: | SOLID-STATE NMR | | Cite: | Conformational Plasticity in Ion Channel Recognition of a Peptide Toxin
To be Published
|
|
4V70
 
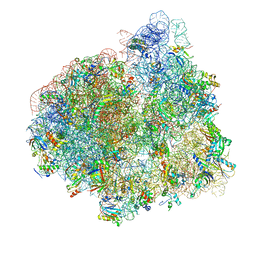 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8GY5
 
 | |
2UW3
 
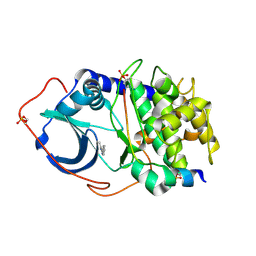 | | Structure of PKA-PKB chimera complexed with 5-methyl-4-phenyl-1H- pyrazole | | Descriptor: | 3-METHYL-4-PHENYL-1H-PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
8H62
 
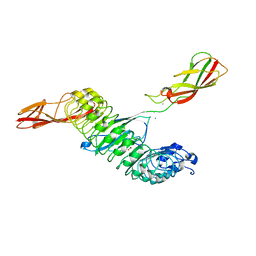 | | Crystal structure of Internalin A from Listeria monocytogenes with human E-cadherin EC12 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-1, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
4UZM
 
 | |
