9ETU
 
 | | Archaellum filament from the Halobacterium salinarum deltaAgl27 strain | | Descriptor: | 3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
8SHT
 
 | | CCT G beta 5 complex closed state 14 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SH9
 
 | | CCT G beta 5 complex best PhLP1 class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SG9
 
 | | CCT G beta 5 complex closed state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8T20
 
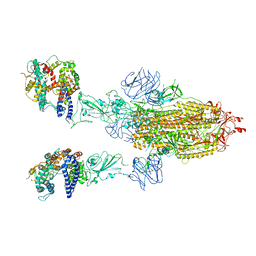 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
5WVI
 
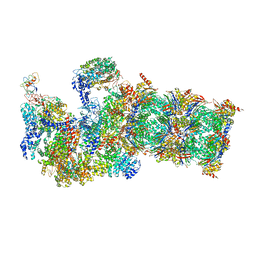 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
8SGC
 
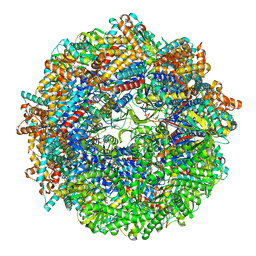 | | CCT G beta 5 complex closed state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SFF
 
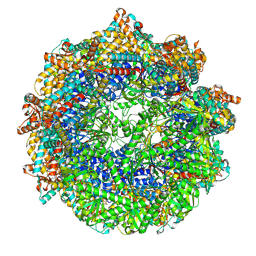 | | CCT G beta 5 complex closed state 0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHN
 
 | | CCT-G beta 5 complex closed state 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SGQ
 
 | | CCT G beta 5 complex intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SUZ
 
 | | Open State of the SARS-CoV-2 Envelope Protein Transmembrane Domain, Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Medeiros-Silva, J, Dregni, A.J, Somberg, N.H, Hong, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure of the open SARS-CoV-2 E viroporin.
Sci Adv, 9, 2023
|
|
8SHG
 
 | | CCT G beta 5 complex closed state 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
4KPS
 
 | | Structure and receptor binding specificity of the hemagglutinin H13 from avian influenza A virus H13N6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Lu, X, Qi, J, Shi, Y, Gao, G. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structure and Receptor Binding Specificity of Hemagglutinin H13 from Avian Influenza A Virus H13N6
J.Virol., 87, 2013
|
|
8SG8
 
 | | CCT G beta 5 complex closed state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHQ
 
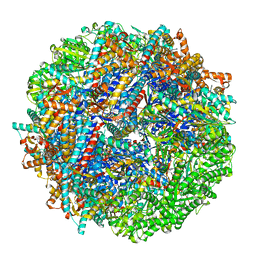 | | CCT G beta 5 complex closed state 12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHF
 
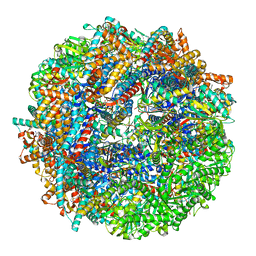 | | CCT-G beta 5 complex closed state 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SWZ
 
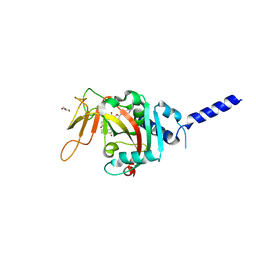 | | PARP4 ART domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
6DXX
 
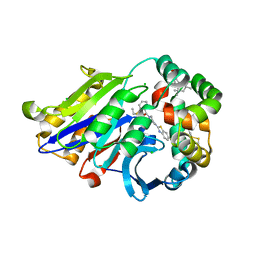 | | Human N-acylethanolamine-hydrolyzing acid amidase (NAAA) in complex with non-covalent benzothiazole-piperazine inhibitor ARN19702, in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8SLZ
 
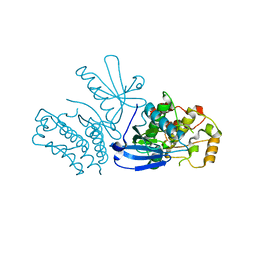 | |
8SX1
 
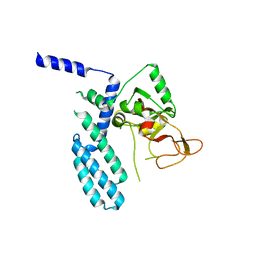 | | PARP4 catalytic domain | | Descriptor: | Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
4KSZ
 
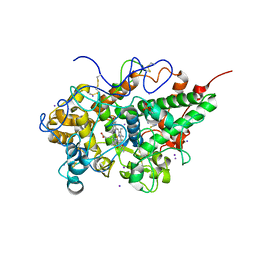 | | Crystal structure of bovine lactoperoxidase complexed with cystiene at 1.98A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, R.P, Singh, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of bovine lactoperoxidase complexed with cystiene at 1.98A resolution
To be Published
|
|
8SX2
 
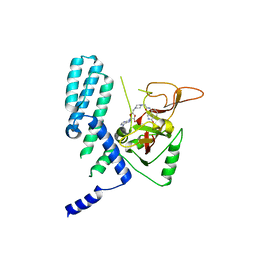 | | PARP4 catalytic domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SWY
 
 | | PARP4 ART domain bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8T1P
 
 | |
