5LQS
 
 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
3QEZ
 
 | | Crystal structure of the mutant T159V,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5431 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMS
 
 | | Crystal structure of the mutant T159V,V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QW4
 
 | | Structure of Leishmania donovani UMP synthase | | Descriptor: | CHLORIDE ION, UMP synthase, URIDINE-5'-MONOPHOSPHATE | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3RLU
 
 | | Crystal structure of the mutant K82A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QF0
 
 | | Crystal structure of the mutant T159V,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMR
 
 | | Crystal structure of the mutant R160A,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3213 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QMT
 
 | | Crystal structure of the mutant V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3202 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3RLV
 
 | | Crystal structure of the mutant Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3P60
 
 | | Crystal structure of the mutant T159V of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3P5Y
 
 | | Crystal structure of the mutant T159A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3TQ0
 
 | | Crystal structure of Leishmania major dihydroorotate dehydrogenase in complex with fumarate | | Descriptor: | Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Cordeiro, A.T, Feliciano, P.R, Nonato, M.C. | | Deposit date: | 2011-09-08 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydroorotate dehydrogenase from Leishmania major.
Biochimie, 94, 2012
|
|
3SJ3
 
 | | Crystal structure of the mutant R160A.Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3V1P
 
 | | Crystal structure of the mutant Q185A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-12-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3VI2
 
 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
6I0R
 
 | | Structure of quinolinate synthase in complex with 5-mercaptopyridine-2,3-dicarboxylic acid | | Descriptor: | 5-mercaptopyridine-2,3-dicarboxylic acid, FE (III) ION, HYDROSULFURIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
8FVC
 
 | | E coli. CTP synthase in complex with dF-dCTP (potassium malonate + 5 mM MgCl2) | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVA
 
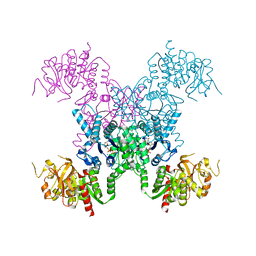 | | E coli. CTP synthase in complex with F-araCTP | | Descriptor: | 4-amino-1-{2-deoxy-2-fluoro-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, CTP synthase, MALONIC ACID, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVD
 
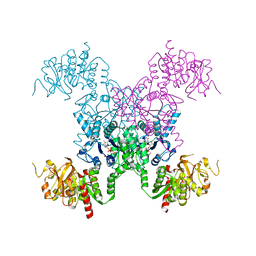 | | E coli. CTP synthase in complex with dF-dCTP (potassium malonate + 100 mM MgCl2) | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MAGNESIUM ION, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV7
 
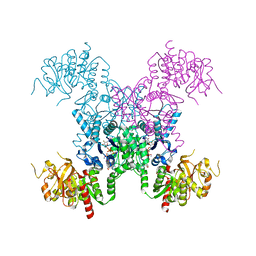 | | E coli. CTP synthase in complex with dF-dCTP + ATP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
6X11
 
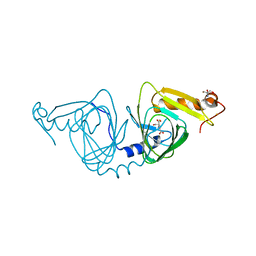 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - an enol tautomer of ACMS monodentately bound structure | | Descriptor: | (2Z,3Z)-2-[(2Z)-3-hydroxyprop-2-en-1-ylidene]-3-iminobutanedioic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-05-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I0P
 
 | | Structure of quinolinate synthase in complex with 6-mercaptopyridine-2,3-dicarboxylic acid | | Descriptor: | 6-mercaptopyridine-2,3-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
6I0K
 
 | | Structure of quinolinate synthase in complex with 4-mercaptophthalic acid | | Descriptor: | 4-mercaptoidenecyclohexa-2,5-diene-1,2-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3NVA
 
 | | Dimeric form of CTP synthase from Sulfolobus solfataricus | | Descriptor: | CTP synthase | | Authors: | Harris, P, Willemoes, M, Lauritsen, I, Johansson, E, Jensen, K.F. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structure of the dimeric form of CTP synthase from Sulfolobus solfataricus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
