5MPA
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8BY4
 
 | |
1F98
 
 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT T50V | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-07 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
6UJY
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (-)-3TC-TP | | Descriptor: | Lamivudine Triphosphate, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59456086 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
7B6G
 
 | | Crystal structure of MurE from E.coli in complex with Z1675346324 | | Descriptor: | DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, trans-3-[(2,6-dimethylpyrimidin-4-yl)(methyl)amino]cyclobutan-1-ol | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
1TWX
 
 | | Crystal structure of the thrombin mutant D221A/D222K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, Prothrombin | | Authors: | Pineda, A.O, Zhang, E, Guinto, E.R, Savvides, S.N, Tulinsky, A, Di Cera, E. | | Deposit date: | 2004-07-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the thrombin mutant D221A/D222K: the Asp222:Arg187 ion-pair stabilizes the fast form
Biophys.Chem., 112, 2004
|
|
7B6Q
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | ISOPROPYL ALCOHOL, N-[(furan-2-yl)methyl]-1H-benzimidazol-2-amine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6UJZ
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (+)-FTC-TP | | Descriptor: | MAGNESIUM ION, SULFATE ION, [[(2~{S},5~{R})-5-(4-azanyl-5-fluoranyl-2-oxidanylidene-pyrimidin-1-yl)-1,3-oxathiolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55642557 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
1WUY
 
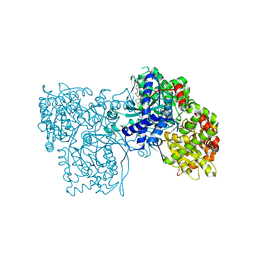 | | Crystallographic studies on acyl ureas, a new class of inhibitors of glycogen phosphorylase. Broad specificity of the allosteric site | | Descriptor: | 4-[3-CHLORO-4-({[(2,4-DICHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]BUTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Klabunde, T, Wendt, K.U, Defossa, E. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
7B68
 
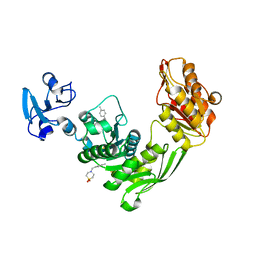 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | 4-[(4-methylphenyl)methyl]-1,4-thiazinane 1,1-dioxide, DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6UK0
 
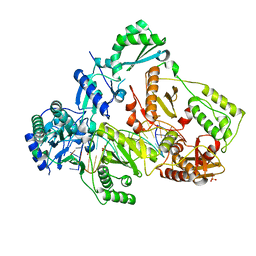 | | HIV-1 M184V reverse transcriptase-DNA complex | | Descriptor: | MAGNESIUM ION, Primer DNA, SULFATE ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75695229 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
8TLY
 
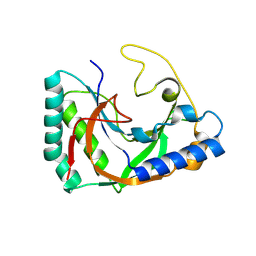 | |
1FAO
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-13 | | Release date: | 2000-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
5MPC
 
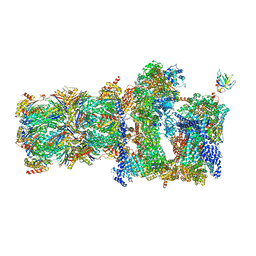 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7B61
 
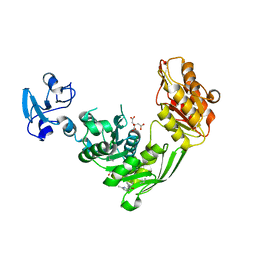 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | (R)-N-(1-cyclopropylethyl)-6-methylpicolinamide, (S)-N-(1-cyclopropylethyl)-6-methylpicolinamide, CITRIC ACID, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6O
 
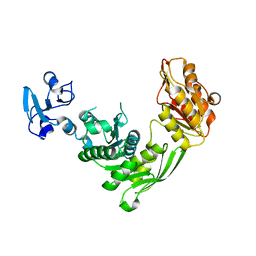 | | Crystal structure of E.coli MurE mutant - C269S C340S C450S | | Descriptor: | ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
4XRS
 
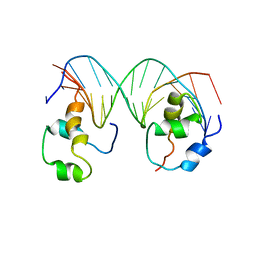 | | Heterodimeric complex of transcription factors MEIS1 and DLX3 on specific DNA | | Descriptor: | DNA (5'-D(P*AP*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*GP*AP*TP*AP*AP*TP*TP*GP*TP*T)-3'), ... | | Authors: | Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Morgunova, E, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
6ULM
 
 | | Crystal structure of human cadherin 17 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-17 | | Authors: | Gray, M.E, Sotomayor, M. | | Deposit date: | 2019-10-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the nonclassical cadherin-17 N-terminus and implications for its adhesive binding mechanism.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8FJ5
 
 | | Structure of the Haloferax volcanii archaeal type IV pilus | | Descriptor: | Pilin_N domain-containing protein | | Authors: | Wang, F, Kreutzberger, M.A, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FJS
 
 | |
5MVA
 
 | |
8FK7
 
 | | Structure of the Pyrobaculum calidifontis flagellar-like archaeal type IV pilus | | Descriptor: | Flagellin | | Authors: | Wang, F, Kreutzberger, M.A, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5MPP
 
 | | Structure of AaLS-wt | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
7B6K
 
 | | Crystal structure of MurE from E.coli in complex with Z57715447 | | Descriptor: | 5-cyclohexyl-3-(pyridin-4-yl)-1,2,4-oxadiazole, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
2IH2
 
 | |
