6A1Z
 
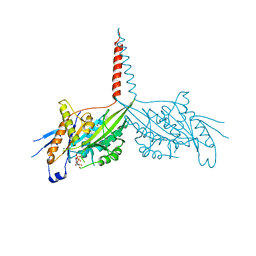 | | Crystal Structure of dimeric Kinesin-3 KIF13B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin family member 13B, MAGNESIUM ION | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-06-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A2D
 
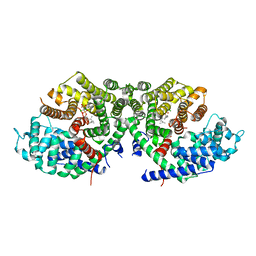 | | Crystal structure of a synthase 2 from santalum album in complex with ligand1 | | Descriptor: | MAGNESIUM ION, Sesquisabinene B synthase 2, [bis(chloranyl)-[oxidanyl-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienoxy]phosphoryl]methyl]phosphonic acid | | Authors: | Han, X, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-10 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a synthase 2 from santalum album
To be published
|
|
5ZWP
 
 | |
8HP8
 
 | |
4U5B
 
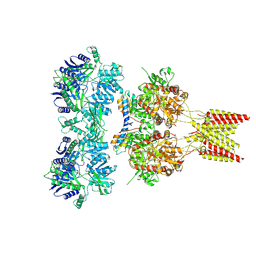 | | Crystal structure of GluA2 A622T, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5037 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U5F
 
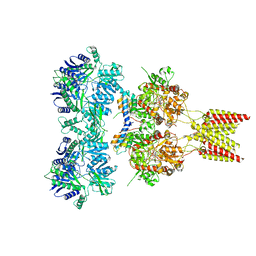 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex, GluA2cryst2 construct | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
8HAM
 
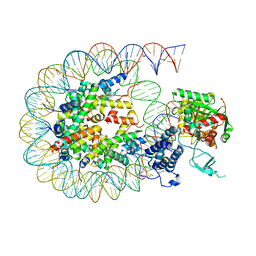 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
6A39
 
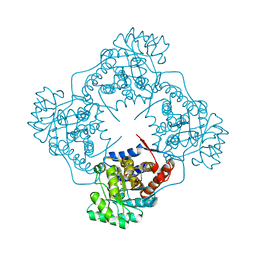 | | The crystal structure of Mandelate oxidase Y128F with C4a-Malic acid-monooxide-FMN adduct | | Descriptor: | (~{E})-2-[[(4~{a}~{S})-7,8-dimethyl-2,4-bis(oxidanylidene)-10-[(2~{S},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-5~{H}-benzo[g]pteridin-4~{a}-yl]oxy]-3-oxidanyl-but-2-enedioic acid, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The crystal structure of Mandelate oxidase mutant Y128F with C4a-Malic acid-monooxide-FMN adduct
To Be Published
|
|
7B14
 
 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
8HAN
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
5ZZM
 
 | |
8HKC
 
 | | Cryo-EM structure of E. coli RNAP sigma32 complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wu, S, Ma, L.X. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural Insight into the Mechanism of sigma 32-Mediated Transcription Initiation of Bacterial RNA Polymerase.
Biomolecules, 13, 2023
|
|
7B62
 
 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
5HNC
 
 | | Synchrotron X-ray single crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Kusaka, K, Baba, S, Mizuno, N, Kimura, T. | | Deposit date: | 2016-01-18 | | Release date: | 2016-07-20 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Neutron and X-ray single-crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6A06
 
 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
4U60
 
 | |
8HAH
 
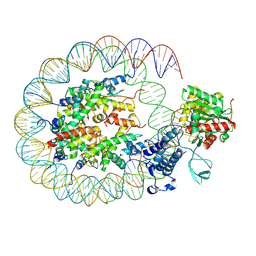 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
6A46
 
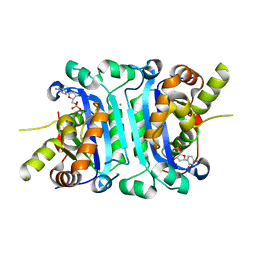 | | Structure of TREX2 in complex with a nucleotide (dCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
8HJE
 
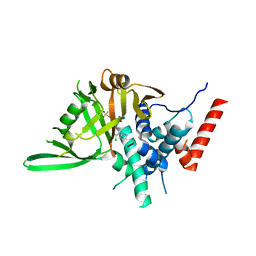 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
5HNL
 
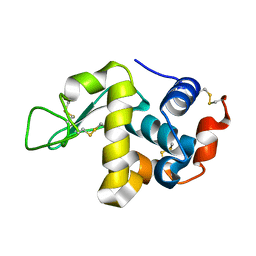 | | In-house X-ray single crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Kusaka, K, Baba, S, Mizuno, N, Kimura, T. | | Deposit date: | 2016-01-18 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Neutron and X-ray single-crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6A4R
 
 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | Descriptor: | ASPARTIC ACID, Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
6A0R
 
 | | Homoserine dehydrogenase from Thermus thermophilus HB8 unliganded form | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
6A19
 
 | | Mandelate oxidase mutant-Y128F with Benzoylformic acid | | Descriptor: | 4-hydroxymandelate oxidase, BENZOYL-FORMIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
