6TA4
 
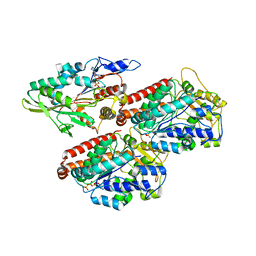 | | Human kinesin-5 motor domain in the AMPPNP state bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A.P, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
1O83
 
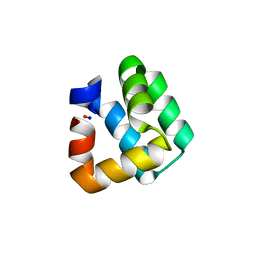 | | Crystal Structure of Bacteriocin AS-48 at pH 7.5, phosphate bound. Crystal form I | | Descriptor: | GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, PHOSPHATE ION | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Valdivia, E, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-25 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
6T7C
 
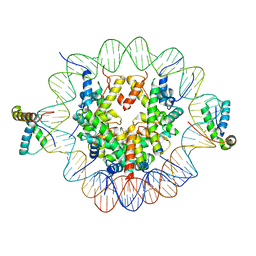 | | Structure of two copies of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6TG4
 
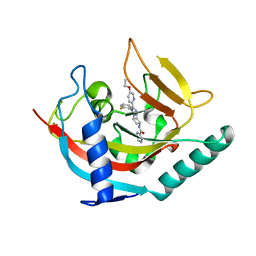 | | Tankyrase 2 in complex with an inhibitor (OM-1700) | | Descriptor: | N-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]pyridine-2-carboxamide, Tankyrase-2, ZINC ION | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
4YJ5
 
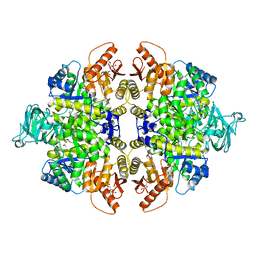 | | Crystal structure of PKM2 mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
6T7A
 
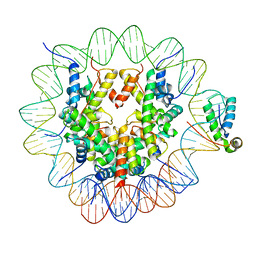 | | Structure of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6T7B
 
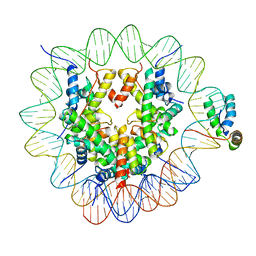 | | Structure of human Sox2 transcription factor in complex with a nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
4YUB
 
 | |
6TKQ
 
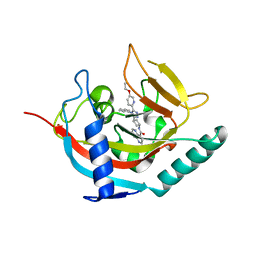 | | Tankyrase 2 in complex with an inhibitor (OM-2700) | | Descriptor: | Tankyrase-2, ZINC ION, ~{N}-[3-[5-(5-ethoxypyridin-2-yl)-4-phenyl-1,2,4-triazol-3-yl]cyclobutyl]-1,5-naphthyridine-4-carboxamide | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
6TCA
 
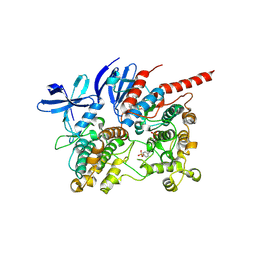 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
4YPU
 
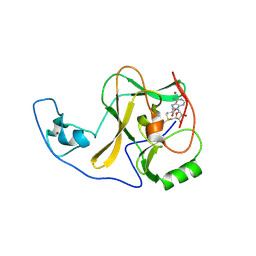 | | ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
7TRB
 
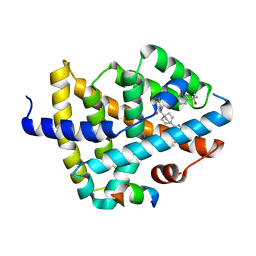 | | CRYSTAL STRUCTURE OF FARNESOID X-ACTIVATED RECEPTOR COMPLEXED WITH COMPOUND-32 AKA (1S,3S)-N-({4-[5-(2-FLUOROPR OPAN-2-YL)-1,2,4-OXADIAZOL-3-YL]BICYCLO[2.2.2]OCTAN-1-YL}M ETHYL)-3-HYDROXY-N-[4'-(2-HYDROXYPROPAN-2-YL)-[1,1'-BIPHEN YL]-3-YL]-3-(TRIFLUOROMETHYL)CYCLOBUTANE-1-CARBOXAMIDE | | Descriptor: | (1s,3s)-N-({4-[5-(2-fluoropropan-2-yl)-1,2,4-oxadiazol-3-yl]bicyclo[2.2.2]octan-1-yl}methyl)-3-hydroxy-N-[4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]-3-(trifluoromethyl)cyclobutane-1-carboxamide, Bile acid receptor, co-activator | | Authors: | Khan, J.A, Ruzanov, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
7TY3
 
 | | Crystal Structure of SETD2 Bound to an Indole-based Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETD2, N-[(1R,3R)-3-(4-acetylpiperazin-1-yl)cyclohexyl]-4-fluoro-7-methyl-1H-indole-2-carboxamide, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational-Design-Driven Discovery of EZM0414: A Selective, Potent SETD2 Inhibitor for Clinical Studies.
Acs Med.Chem.Lett., 13, 2022
|
|
7TY2
 
 | | Crystal Structure of SETD2 Bound to an Indole-based Inhibitor | | Descriptor: | Histone-lysine N-methyltransferase SETD2, N-[(1R,3S)-3-(4-acetylpiperazin-1-yl)cyclohexyl]-4-fluoro-7-methyl-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Conformational-Design-Driven Discovery of EZM0414: A Selective, Potent SETD2 Inhibitor for Clinical Studies.
Acs Med.Chem.Lett., 13, 2022
|
|
6B42
 
 | |
1OW8
 
 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
6CBI
 
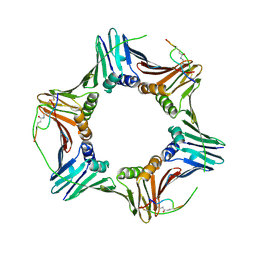 | | PCNA in complex with inhibitor | | Descriptor: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Bruning, J.B, Wegener, K.L. | | Deposit date: | 2018-02-03 | | Release date: | 2018-07-04 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
6CCD
 
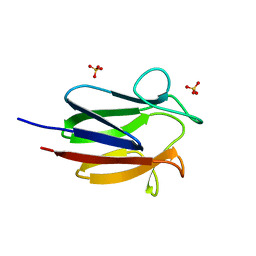 | |
6CF6
 
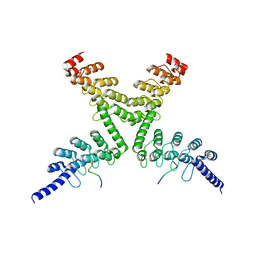 | | RNF146 TBM-Tankyrase ARC2-3 complex | | Descriptor: | RNF146, Tankyrase-1 | | Authors: | Da Rosa, P.A, Xu, W. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
Protein Sci., 27, 2018
|
|
6T9L
 
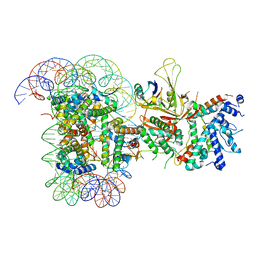 | |
6TA3
 
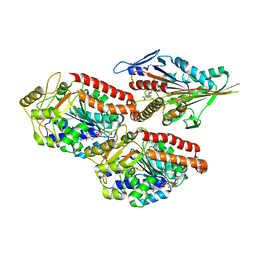 | | Human kinesin-5 motor domain in the GSK-1 state bound to microtubules (Conformation 1) | | Descriptor: | 6-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-quinolin-2-one, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
1MQZ
 
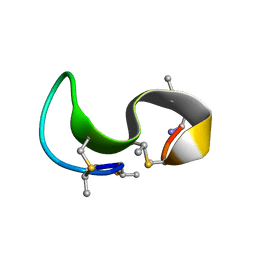 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
1MQY
 
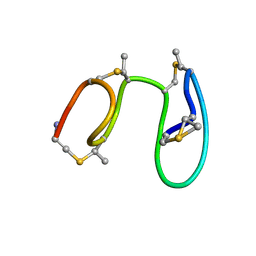 | | NMR solution structure of type-B lantibiotics mersacidin in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
6CPI
 
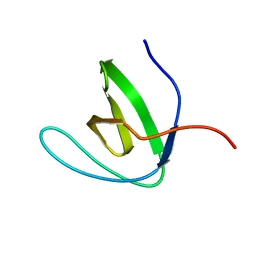 | | Solution structure of SH3 domain from Shank1 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
