7AS4
 
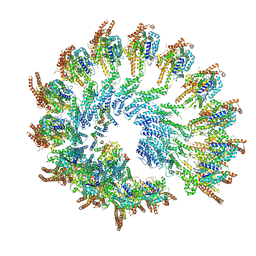 | | Recombinant human gTuRC | | Descriptor: | Actin, cytoplasmic 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Serna, M, Fernandez-Leiro, R, Llorca, O. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Assembly of the asymmetric human gamma-tubulin ring complex by RUVBL1-RUVBL2 AAA ATPase.
Sci Adv, 6, 2020
|
|
7E1W
 
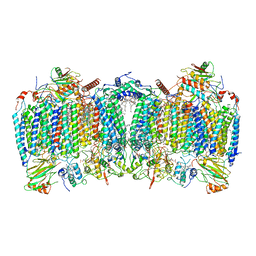 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
4HI4
 
 | |
7AJC
 
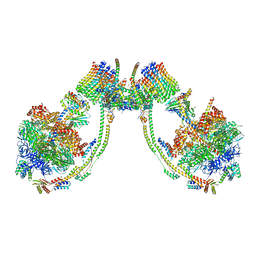 | | bovine ATP synthase dimer state1:state2 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5F5K
 
 | |
7E8D
 
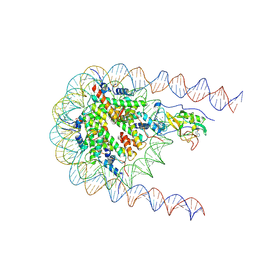 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7E9T
 
 | | Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhu, Y, Tai, L, Zhu, G, Yin, G, Sun, F. | | Deposit date: | 2021-03-05 | | Release date: | 2021-11-17 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AFT
 
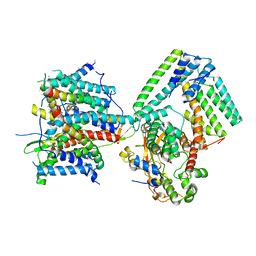 | | Cryo-EM structure of the signal sequence-engaged post-translational Sec translocon | | Descriptor: | Mating factor alpha-1,Mating factor alpha-1, Protein translocation protein SEC63, Protein transport protein SBH1, ... | | Authors: | Weng, T.-H, Beatrix, B, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the active post-translational Sec translocon.
Embo J., 40, 2021
|
|
4HKO
 
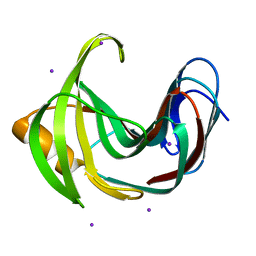 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7AJG
 
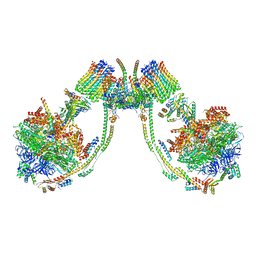 | | bovine ATP synthase dimer state2:state3 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4HKW
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with Substrate and Products | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endo-1,4-beta-xylanase 2, ... | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P, Coates, L. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7PDV
 
 | |
7AQA
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H382A | | Descriptor: | (dicuprio-$l^{3}-sulfanyl)-sulfanyl-copper, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
5EXT
 
 | |
7AIJ
 
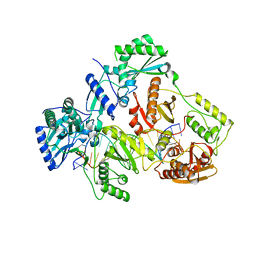 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AQC
 
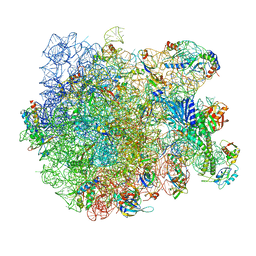 | | Structure of the bacterial RQC complex (Decoding State) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Mimicry of Canonical Translation Elongation Underlies Alanine Tail Synthesis in RQC.
Mol.Cell, 81, 2021
|
|
8OJP
 
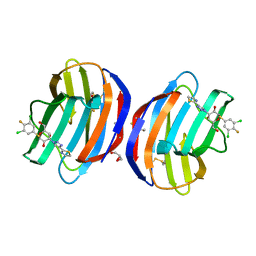 | | Human galectin 1 in complex with inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-[3,5-bis(chloranyl)-4-fluoranyl-phenyl]sulfanyl-6-(hydroxymethyl)-4-[4-(1,3-thiazol-2-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, GLYCEROL, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Nilsson, U.J, Zetterberg, F.R, Peterson, K. | | Deposit date: | 2023-03-24 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of Selective and Orally Available Galectin-1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8OUO
 
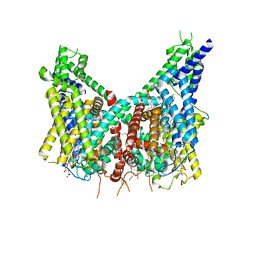 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 32, 2024
|
|
7AOK
 
 | | Crystal structure of CI2 mutant L49I | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
7DYW
 
 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DY4
 
 | |
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYX
 
 | | Human JMJD5 in complex with MN and 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7AS9
 
 | |
7DYU
 
 | | Human JMJD5 in complex with MN and 5-((4-phenylbutyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(4-phenylbutylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
