3KGB
 
 | |
6G3W
 
 | |
6GAJ
 
 | |
4P6D
 
 | | Structure of ribB complexed with PO4 ion | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
9EDV
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1e: 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-guided optimization of small molecules targeting Yck2 as a strategy to combat Candida albicans.
Nat Commun, 16, 2025
|
|
9EDY
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2b: 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine | | Descriptor: | 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided optimization of small molecules targeting Yck2 as a strategy to combat Candida albicans.
Nat Commun, 16, 2025
|
|
9EDX
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2a: 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-guided optimization of small molecules targeting Yck2 as a strategy to combat Candida albicans.
Nat Commun, 16, 2025
|
|
9EDW
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1f: 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine | | Descriptor: | 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine, CHLORIDE ION, Non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-guided optimization of small molecules targeting Yck2 as a strategy to combat Candida albicans.
Nat Commun, 16, 2025
|
|
6RWG
 
 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
3WTJ
 
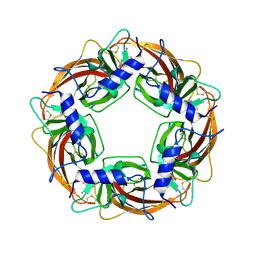 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Thiacloprid | | Descriptor: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
7LT8
 
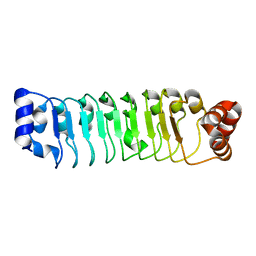 | | Crystal structure of Ras suppressor-1 | | Descriptor: | Ras suppressor protein 1 | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2021-02-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76000249 Å) | | Cite: | Molecular basis for Ras suppressor-1 binding to PINCH-1 in focal adhesion assembly.
J.Biol.Chem., 296, 2021
|
|
7LT9
 
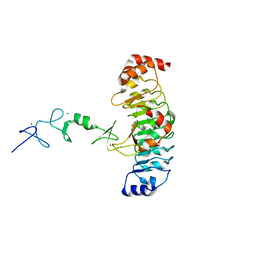 | |
7M8K
 
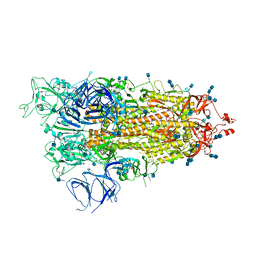 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
5CGA
 
 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(1,3,5-trimethyl-1H-pyrazole-4-yl)ethanol | | Descriptor: | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
2QEZ
 
 | |
5DHZ
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5G5K
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
3KK2
 
 | | HIV-1 reverse transcriptase-DNA complex with dATP bound in the nucleotide binding site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*A*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', 5'-D(*AP*CP*A*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
6GWQ
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with an Inhibitory Nanobody (VHH-2g-42) | | Descriptor: | Plasminogen Activator Inhibitor-1, VHH-2g-42 | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2018-06-25 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
6SEU
 
 | | X-ray structure of the gold/lysozyme adduct formed upon 21h exposure of protein crystals to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Lysozyme C, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
6KK8
 
 | | XN joint refinement of manganese catalase from Thermus Thermophilus HB27 | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (III) ION, OXYGEN ATOM, ... | | Authors: | Yamada, T, Yano, N, Kusaka, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.37 Å), X-RAY DIFFRACTION | | Cite: | Single-crystal time-of-flight neutron Laue methods: application to manganese catalase from Thermus thermophilus HB27
J.Appl.Crystallogr., 2019
|
|
6GWP
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with Two Inhibitory Nanobodies (VHH-2g-42, VHH-2w-64) | | Descriptor: | Plasminogen Activator Inhibitor-1, VHH-2g-42, VHH-2w-64 | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2018-06-25 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
3L1V
 
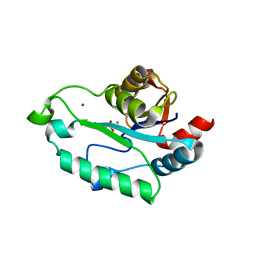 | | Crystal structure of GmhB from E. coli in complex with calcium and phosphate. | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, PHOSPHATE ION, ... | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
7A3O
 
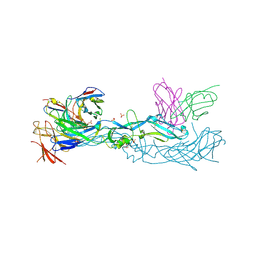 | | Crystal structure of dengue 1 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | Core protein, GLYCEROL, SULFATE ION, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
