5ZO3
 
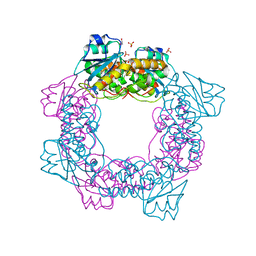 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
1A1V
 
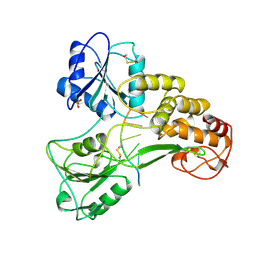 | | HEPATITIS C VIRUS NS3 HELICASE DOMAIN COMPLEXED WITH SINGLE STRANDED SDNA | | Descriptor: | DNA (5'-D(*UP*UP*UP*UP*UP*UP*UP*U)-3'), PROTEIN (NS3 PROTEIN), SULFATE ION | | Authors: | Kim, J.L, Morgenstern, K.A, Griffith, J.P, Dwyer, M.D, Thomson, J.A, Murcko, M.A, Lin, C, Caron, P.R. | | Deposit date: | 1997-12-17 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding.
Structure, 6, 1998
|
|
5ZO4
 
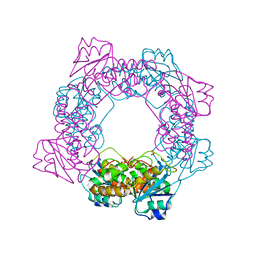 | | inactive state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO5
 
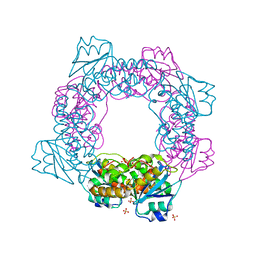 | | active state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
1HS1
 
 | |
1HS3
 
 | |
1HS8
 
 | |
1HS4
 
 | |
3D84
 
 | |
2F1L
 
 | |
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
4QJF
 
 | |
5N6F
 
 | | Crystal structure of TGT in complex with guanine fragment | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GUANINE, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2017-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.11909521 Å) | | Cite: | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
5DHB
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5DHC
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
6RKT
 
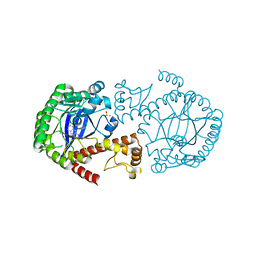 | | Crystal Structure of TGT in complex with N2-methyl-1H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
6RKQ
 
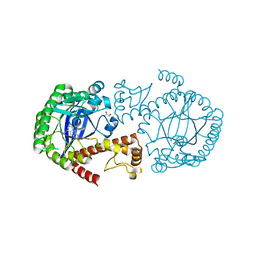 | | Crystal Structure of TGT in complex with N2-methyl-8-(prop-1-yn-1-yl)-3H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | (8~{R})-~{N}2-methyl-8-prop-1-ynyl-7,8-dihydro-3~{H}-imidazo[4,5-g]quinazoline-2,6-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
2HOL
 
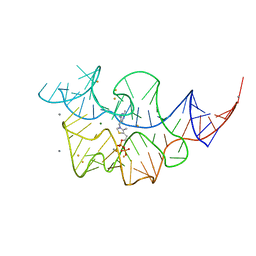 | |
2HOJ
 
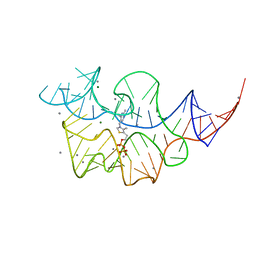 | |
2HOP
 
 | |
2P4V
 
 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
2HOO
 
 | |
4OXP
 
 | |
2FCA
 
 | | The structure of BsTrmB | | Descriptor: | POTASSIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zegers, I, Van Vliet, F, Bujnicki, J, Kosinski, J, Gigot, D, Droogmans, L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase.
Nucleic Acids Res., 34, 2006
|
|
