3K4Y
 
 | |
3K56
 
 | |
3K5F
 
 | | Human BACE-1 COMPLEX WITH AYH011 | | Descriptor: | (1R,3S)-3-[1-(acetylamino)-1-methylethyl]-N-[(1S,2S,4R)-1-benzyl-5-(butylamino)-2-hydroxy-4-methyl-5-oxopentyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
3JRE
 
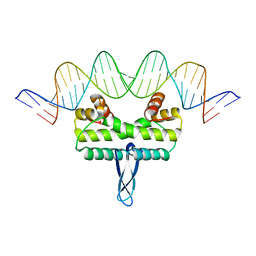 | |
3JSA
 
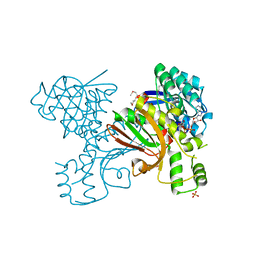 | | Homoserine dehydrogenase from Thermoplasma volcanium complexed with NAD | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Osipiuk, J, Nocek, B, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of Homoserine dehydrogenase from Thermoplasma volcanium
To be Published
|
|
3K14
 
 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
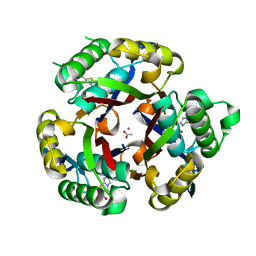
3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
3K42
 
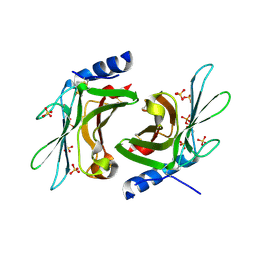 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-dependent mannose-6-phosphate receptor, SN-GLYCEROL-1-PHOSPHATE, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K77
 
 | | X-ray crystal structure of XRCC1 | | Descriptor: | DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, London, R.E. | | Deposit date: | 2009-10-12 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JTL
 
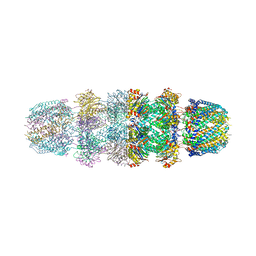 | |
3JVL
 
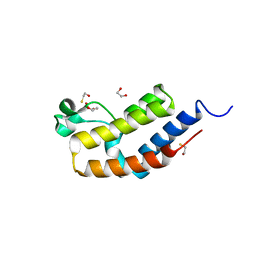 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3JW6
 
 | |
3K06
 
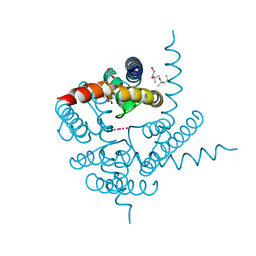 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4WHD
 
 | | Human CEACAM1 N-domain homodimer | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Kirouac, K.N, Prive, G.G. | | Deposit date: | 2014-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human CEACAM1 N-domain homodimer
To Be Published
|
|
3K0G
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K86
 
 | |
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
3K9J
 
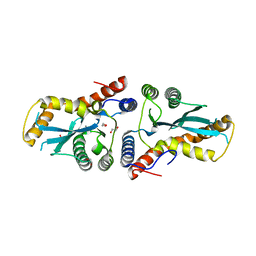 | | Transposase domain of Metnase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Histone-lysine N-methyltransferase SETMAR | | Authors: | Goodwin, K.D, He, H, Imasaki, T, Lee, S.-H, Georgiadis, M.M. | | Deposit date: | 2009-10-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase.
Biochemistry, 49, 2010
|
|
3JV4
 
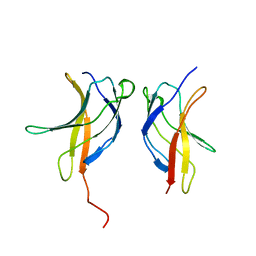 | |
3JZ9
 
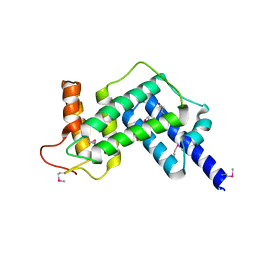 | | Crystal structure of the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3K01
 
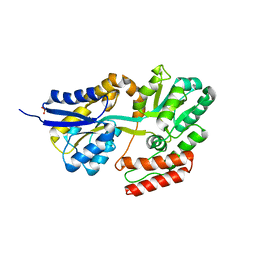 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
3K1N
 
 | |
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
3JVK
 
 | | Crystal structure of bromodomain 1 of mouse Brd4 in complex with histone H3-K(ac)14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, histone H3.3 peptide | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
