8CMZ
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8HG3
 
 | | Cryo-EM structure of the Lhcp complex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4~{S})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-11-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
6GOM
 
 | | KRAS-169 Q61H GPPNHP + PPIN-1 | | Descriptor: | (6~{S})-1-(1~{H}-imidazol-4-ylcarbonyl)-6-[(4-phenylphenyl)methyl]-4-propyl-1,4-diazepan-5-one, CITRIC ACID, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5LFI
 
 | | lactococcin A immunity protein | | Descriptor: | Lactococcin-A immunity protein | | Authors: | Persson, C, Fuochi, V, Pedersen, A, Karlsson, B.G, Nissen-Meyer, J, Kristiansen, P.E, Oppegard, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure and Mutational Analysis of the Lactococcin A Immunity Protein.
Biochemistry, 55, 2016
|
|
6N5W
 
 | |
6YCY
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
6WFN
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6OM5
 
 | | Structure of a haemophore from Haemophilus haemolyticus | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Torrado, M, Walshe, J.L, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A heme-binding protein produced by Haemophilus haemolyticus inhibits non-typeable Haemophilus influenzae.
Mol.Microbiol., 113, 2020
|
|
8FIX
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
6O30
 
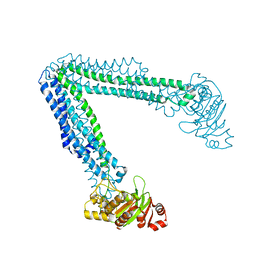 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
8F2K
 
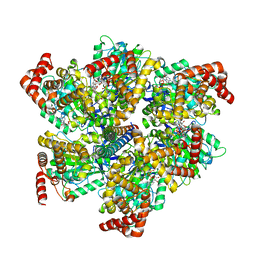 | |
9C87
 
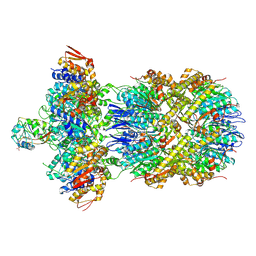 | | Cryo-EM Structure of a Proteolytic ClpXP AAA+ Machine Poised to Unfold a Linear-Degron DHFR-ssrA Substrate Bound with MTX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Sauer, R.T, Davis, J.H. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A proteolytic AAA+ machine poised to unfold protein substrates.
Nat Commun, 15, 2024
|
|
6YQO
 
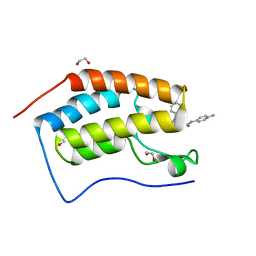 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | Descriptor: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6YCZ
 
 | | Plasmodium falciparum Myosin A delta-Nter, Post-Rigor state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
6S25
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with a benzodiazepine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{tert}-butyl ~{N}-[3-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]propyl]carbamate | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with a benzodiazepine ligand
To Be Published
|
|
5G51
 
 | | High resolution structure of the part of VP3 protein of Deformed Wing Virus forming P-domain | | Descriptor: | DWV-VP3-P-DOMAIN | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G52
 
 | | Crystallographic structure of full particle of Deformed Wing Virus | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YYH
 
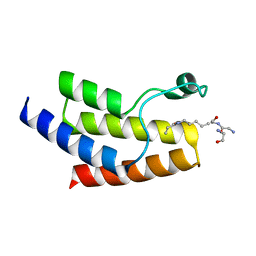 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYN
 
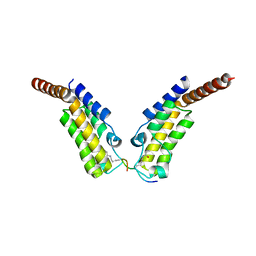 | | Crystal structure of TAF1 BD2 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6OQO
 
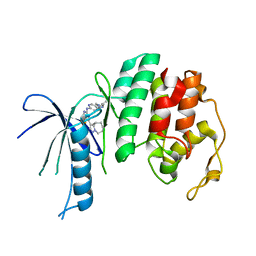 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4YYK
 
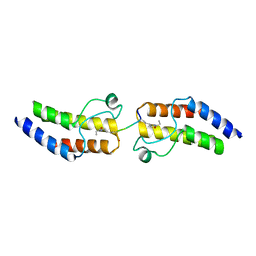 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
8CJU
 
 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CHC
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Deicsics, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CI6
 
 | | PBP AccA from A. vitis S4 in complex with D-glucose-2-phosphate (G2P) | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate binding protein (Agrocinopine) | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
