5GM7
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Ribosome inactivating protein | | Authors: | Singh, B, Singh, P.K, Pandey, S.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-13 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution
To Be Published
|
|
2ODI
 
 | | Restriction Endonuclease BCNI-Cognate DNA Substrate Complex | | Descriptor: | 5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3', 5'-D(*CP*TP*CP*CP*GP*GP*GP*TP*TP*GP*T)-3', CALCIUM ION, ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
2OA9
 
 | | Restriction endonuclease MvaI in the absence of DNA | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kaus-Drobek, M, Czapinska, H, Sokolowska, M, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.
Nucleic Acids Res., 35, 2007
|
|
5EE7
 
 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
2Q10
 
 | | RESTRICTION ENDONUCLEASE BcnI (WILD TYPE)-COGNATE DNA SUBSTRATE COMPLEX | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3'), ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2007-05-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Monomeric Restriction Endonuclease BcnI in the Apo Form and in an Asymmetric Complex with Target DNA.
J.Mol.Biol., 369, 2007
|
|
5E5Q
 
 | | Racemic snakin-1 in P21/c | | Descriptor: | Snakin-1 | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Y
 
 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5EM0
 
 | | Crystal structure of mugwort allergen Art v 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Pollen allergen Art v 4.01, SODIUM ION | | Authors: | Offermann, L.R, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
5EMS
 
 | | Crystal Structure of an iodinated insulin analog | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Lawrence, M.C, Pandyarajan, V, Wan, Z, Weiss, M.A. | | Deposit date: | 2015-11-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extending Halogen-based Medicinal Chemistry to Proteins: IODO-INSULIN AS A CASE STUDY.
J. Biol. Chem., 291, 2016
|
|
5EV0
 
 | | Crystal structure of ragweed profilin Amb a 8 in complex with poly-Pro14 | | Descriptor: | PRO-PRO-PRO-PRO-PRO-PRO-PRO-PRO-PRO, Profilin | | Authors: | Offermann, L.R, He, J.Z, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
4CBY
 
 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(1,3-oxazol-5-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
2PQ5
 
 | | Crystal structure of Dual specificity protein phosphatase 13 (DUSP13) | | Descriptor: | Dual specificity protein phosphatase 13 | | Authors: | Ugochukwu, E, Salah, E, Savitsky, P, Barr, A, Pantic, N, Niesen, F, Burgess-Brown, N, Berridge, G, Bunkoczi, G, Uppenberg, J, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dual specificity protein phosphatase 13 (DUSP13).
To be Published
|
|
2PNB
 
 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
4BCP
 
 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
5FRP
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
4B5D
 
 | | Capitella teleta AChBP in complex with psychonicline (3-((2(S)- Azetidinyl)methoxy)-5-((1S,2R)-2-(2-hydroxyethyl)cyclopropyl)pyridine) | | Descriptor: | 2-[(1R,2S)-2-[5-[[(2S)-azetidin-2-yl]methoxy]pyridin-3-yl]cyclopropyl]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nys, M, Ulens, C. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Insights Into the Structural Determinants Required for High- Affinity Binding of Chiral Cyclopropane-Containing Ligands to Alpha4Beta2-Nicotinic Acetylcholine Receptors: An Integrated Approach to Behaviorally Active Nicotinic Ligands.
J.Med.Chem., 55, 2012
|
|
5FIQ
 
 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
5FXC
 
 | | Crystal structure of glycopeptide 22 in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCOPEPTIDE, SCFV-SM3 | | Authors: | Rojas-Ocariz, V, Companon, I, Aydillo, C, Castro-Lopez, J, Jimenez-Barbero, J, Hurtado-Guerrero, R, Avenoza, A, Zurbano, M.M, Corzana, F, Busto, J.H, Peregrina, J.M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Alpha-S-Glycopeptides Derived from Muc1 with a Flexible and Solvent Exposed Sugar Moiety
J.Org.Chem., 81, 2016
|
|
2P5R
 
 | | Crystal structure of the poplar glutathione peroxidase 5 in the oxidized form | | Descriptor: | CALCIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
5FRS
 
 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN 1, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.073 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
2P5Q
 
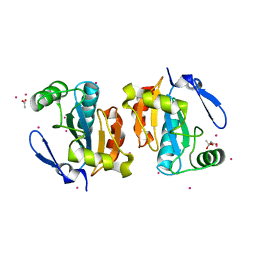 | | Crystal structure of the poplar glutathione peroxidase 5 in the reduced form | | Descriptor: | ACETATE ION, CADMIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
4E02
 
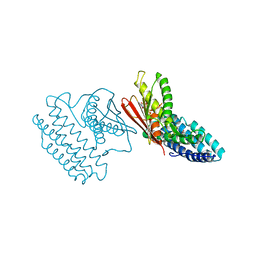 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
5DUQ
 
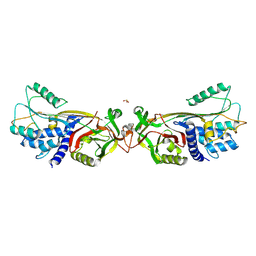 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
2PNA
 
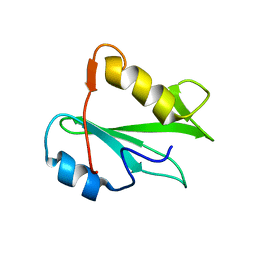 | | STRUCTURE OF AN SH2 DOMAIN OF THE P85 ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL-3-OH KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT N-TERMINAL SH2 DOMAIN | | Authors: | Booker, G.W, Breeze, A.L, Downing, A.K, Panayotou, G, Gout, I, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1992-06-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase.
Nature, 358, 1992
|
|
