8WQR
 
 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
5E5T
 
 | | Quasi-racemic snakin-1 in P1 after radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LKN
 
 | | NMR solution structure of human FNIII domain 2 of NCAM | | Descriptor: | Neural cell adhesion molecule 1 | | Authors: | Slapsak, U, Salzano, G, Amin, L, Abskharon, R.N.N, Ilc, G, Zupancic, B, Biljan, I, Plavec, J, Giachin, G, Legname, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain.
J.Biol.Chem., 291, 2016
|
|
1U08
 
 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
5NZF
 
 | | Complex of H275Y/I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
2O0U
 
 | | Crystal structure of human JNK3 complexed with N-{3-cyano-6-[3-(1-piperidinyl)propanoyl]-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl}-1-naphthalenecarboxamide | | Descriptor: | Mitogen-activated protein kinase 10, N-{3-CYANO-6-[3-(1-PIPERIDINYL)PROPANOYL]-4,5,6,7-TETRAHYDROTHIENO[2,3-C]PYRIDIN-2-YL}1-NAPHTHALENECARBOXAMIDE | | Authors: | Rowland, P, Somers, D. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5NZ4
 
 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZN
 
 | | Complex of H275Y/S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5EE7
 
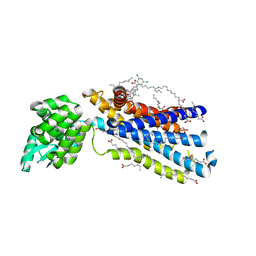 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
8IJR
 
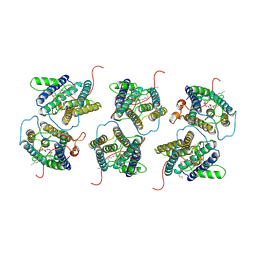 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4GQQ
 
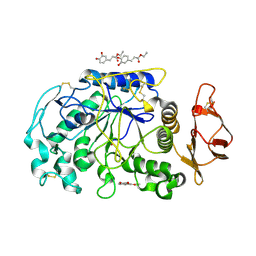 | | Human pancreatic alpha-amylase with bound ethyl caffeate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Williams, L.K, Brayer, G.D. | | Deposit date: | 2012-08-23 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target.
J.Med.Chem., 55, 2012
|
|
5NZE
 
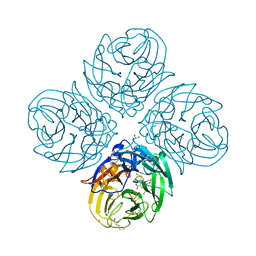 | | Complex of S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Hejdanek, J. | | Deposit date: | 2017-05-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
8GKL
 
 | | Crystal Structure of the Humanized MUC16 Specific Antibody huAR9.6 | | Descriptor: | MUC16 antibody AR9.6 Fab heavy chain, MUC16 antibody AR9.6 Fab light chain, Mucin-16 | | Authors: | Brooks, C.L, Aguilar, E.N. | | Deposit date: | 2023-03-19 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Multivalent MUC16 Recognition and Robust Anti-Pancreatic Cancer Activity of Humanized Antibody AR9.6.
Mol.Cancer Ther., 23, 2024
|
|
8X0S
 
 | | Crystal structure of r(G4C2)2 | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Zhu, G. | | Deposit date: | 2023-11-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Crystal structure of a tetrameric RNA G-quadruplex formed by hexanucleotide repeat expansions of C9orf72 in ALS/FTD.
Nucleic Acids Res., 52, 2024
|
|
5LUD
 
 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
8V98
 
 | | GII.24 Loreto 1972 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
5EM1
 
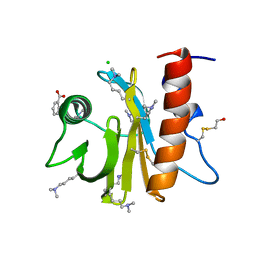 | | Crystal structure of ragweed allergen Amb a 8 | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Profilin | | Authors: | Offermann, L.R, He, J.Z, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
5LLJ
 
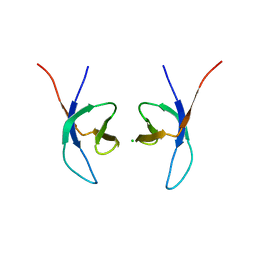 | |
8V9A
 
 | | GII.NA1 Loreto 1257 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Kim, I, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V97
 
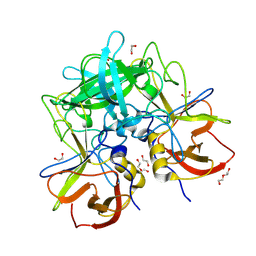 | | GII.17 CS-E1 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, DI(HYDROXYETHYL)ETHER | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
4GQR
 
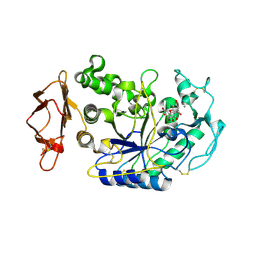 | | Human Pancreatic alpha-amylase in complex with myricetin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CALCIUM ION, ... | | Authors: | Williams, L.K, Brayer, G.D. | | Deposit date: | 2012-08-23 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target.
J.Med.Chem., 55, 2012
|
|
8V95
 
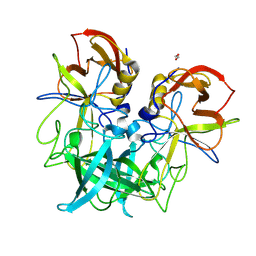 | | GII.8 Amsterdam norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein (Fragment) | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V9Z
 
 | |
1UJ1
 
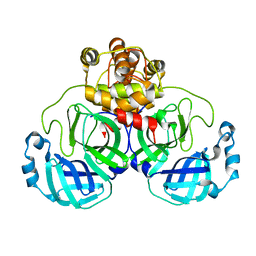 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5NWE
 
 | | Complex of H275Y mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
