1S40
 
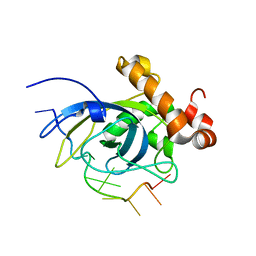 | | SOLUTION STRUCTURE OF THE CDC13 DNA-BINDING DOMAIN COMPLEXED WITH A SINGLE-STRANDED TELOMERIC DNA 11-MER | | Descriptor: | 5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3', Cell division control protein 13 | | Authors: | Mitton-Fry, R.M, Anderson, E.M, Theobald, D.L, Glustrom, L.W, Wuttke, D.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13
J.Mol.Biol., 338, 2004
|
|
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1MBH
 
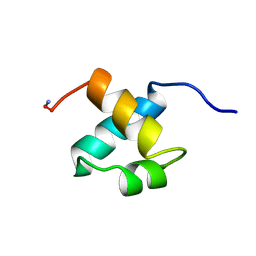 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MSI
 
 | |
1N17
 
 | | Structure and Dynamics of Thioguanine-modified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*(S6G)P*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
1RWU
 
 | |
1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
1S6U
 
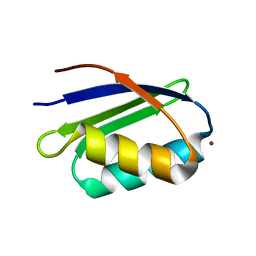 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1RF8
 
 | | Solution structure of the yeast translation initiation factor eIF4E in complex with m7GDP and eIF4GI residues 393 to 490 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic initiation factor 4F subunit p150, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gross, J.D, Moerke, N.J, von der Haar, T, Lugovskoy, A.A, Sachs, A.B, McCarthy, J.E.G, Wagner, G. | | Deposit date: | 2003-11-07 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Cell(Cambridge,Mass.), 115, 2003
|
|
1R7U
 
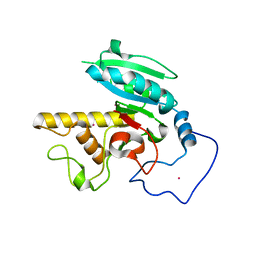 | | Glycosyltransferase B in complex with 3-deoxy-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R82
 
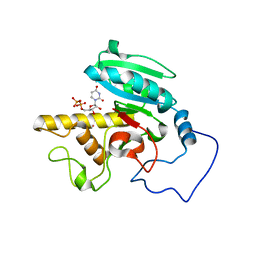 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor, and uridine diphosphate-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1RPC
 
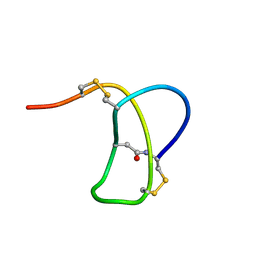 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1PKT
 
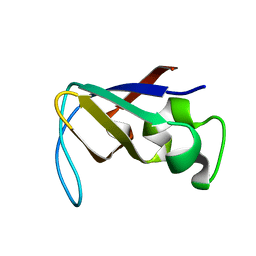 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1TL6
 
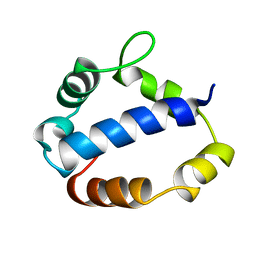 | | Solution structure of T4 bacteriphage AsiA monomer | | Descriptor: | 10 kDa anti-sigma factor | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma70 region 4
Embo J., 23, 2004
|
|
1V4R
 
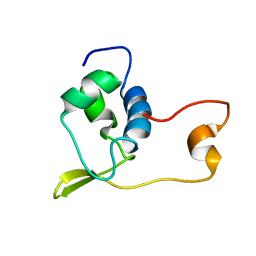 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
1OW8
 
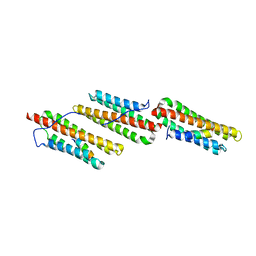 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1P1E
 
 | |
1RT0
 
 | | 12-mer from site II calbindin D9K (DKNGDGEVSFEE) coordinating Zn(II) | | Descriptor: | Vitamin D-dependent calcium-binding protein, intestinal | | Authors: | Ferrari, R, Mendoza, G, James, T, Tonelli, M, Basus, V, Gasque, L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coordination Compounds derived from a dodecapeptide and Pb2+, Cd2+ and Zn2+. Modeling the site II from the Calbindin D9K
To be Published
|
|
1RAU
 
 | |
1RES
 
 | |
1RET
 
 | |
1OWL
 
 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RLP
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RPB
 
 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
