1N14
 
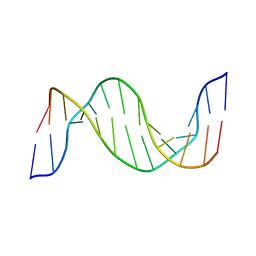 | | Structure and Dynamics of Thioguanine-modified Duplex DNA in Comparison with Unmodified DNA; Structure of Unmodified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*GP*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
1NHN
 
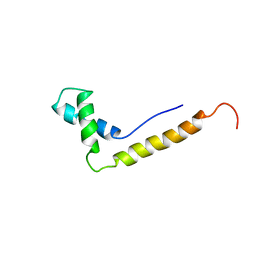 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NHM
 
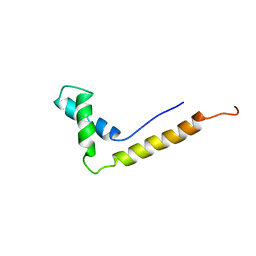 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1POU
 
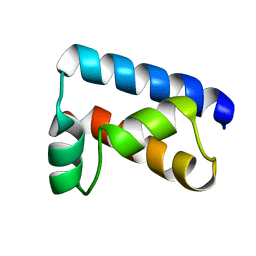 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1Q69
 
 | | Solution structure of T-cell surface glycoprotein CD8 alpha chain and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD8 alpha chain, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1QUZ
 
 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
1QP6
 
 | | SOLUTION STRUCTURE OF ALPHA2D | | Descriptor: | PROTEIN (ALPHA2D) | | Authors: | Hill, R.B, DeGrado, W.F. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha2D, A Nativelike De Novo Designed Protein
J.Am.Chem.Soc., 120, 1998
|
|
1Q8K
 
 | |
1HMJ
 
 | | SOLUTION STRUCTURE OF RNA POLYMERASE SUBUNIT H | | Descriptor: | PROTEIN (SUBUNIT H) | | Authors: | Thiru, A, Hodach, M, Eloranta, J, Kostourou, V, Weinzierl, R. | | Deposit date: | 1999-02-05 | | Release date: | 1999-04-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase subunit H features a beta-ribbon motif within a novel fold that is present in archaea and eukaryotes.
J.Mol.Biol., 287, 1999
|
|
1SRL
 
 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
1HSM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
1HJD
 
 | |
2KQW
 
 | |
1SCL
 
 | |
1HZM
 
 | |
1ICL
 
 | |
1SVJ
 
 | | The solution structure of the nucleotide binding domain of KdpB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-03-29 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
1HZK
 
 | |
1TL6
 
 | | Solution structure of T4 bacteriphage AsiA monomer | | Descriptor: | 10 kDa anti-sigma factor | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma70 region 4
Embo J., 23, 2004
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
1TNQ
 
 | |
1TTY
 
 | | Solution structure of sigma A region 4 from Thermotoga maritima | | Descriptor: | RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
