8RDL
 
 | | Xenorhabdin methyltransferase XrdM with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ToxA protein | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1QOT
 
 | | lectin UEA-II complexed with fucosyllactose and fucosylgalactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 1999-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QZD
 
 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
6IM4
 
 | |
6QMD
 
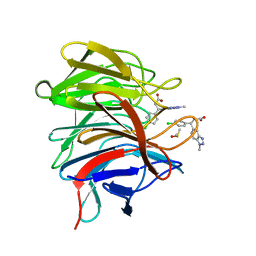 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QME
 
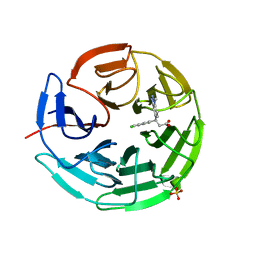 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chloranyl-3-methyl-phenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QMK
 
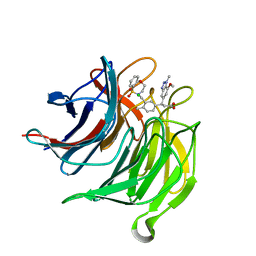 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-[3-[[1,1-bis(oxidanylidene)-3,4-dihydro-5,1$l^{6},2-benzoxathiazepin-2-yl]methyl]-4-methyl-phenyl]-3-(7-methoxy-1-methyl-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QV9
 
 | |
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
6IFO
 
 | | Crystal structure of AcrIIA2-SpyCas9-sgRNA ternary complex | | Descriptor: | AcrIIA2, CRISPR-associated endonuclease Cas9/Csn1, RNA (99-MER) | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race.
Mol. Cell, 73, 2019
|
|
1IG1
 
 | | 1.8A X-Ray structure of ternary complex of a catalytic domain of death-associated protein kinase with ATP analogue and Mn. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, death-associated protein kinase | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-04-16 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
6IG1
 
 | | DNA polymerase IV - DNA ternary complex 10 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN3, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2018-09-21 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
1V2B
 
 | | Crystal Structure of PsbP Protein in the Oxygen-Evolving Complex of Photosystem II from Higher Plants | | Descriptor: | 23-kDa polypeptide of photosystem II oxygen-evolving complex, SULFATE ION, alpha-D-glucopyranose | | Authors: | Ifuku, K, Nakatsu, T, Kato, H, Sato, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-14 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum
Embo Rep., 5, 2004
|
|
7PTQ
 
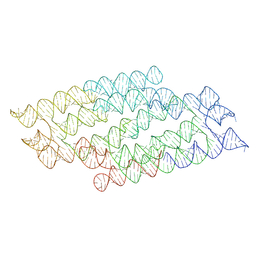 | | RNA origami 5-helix tile | | Descriptor: | Chains: C | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
7PTS
 
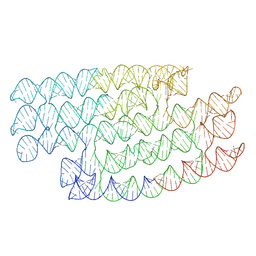 | | RNA origami 5-helix tile | | Descriptor: | 5HT-B | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.71 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
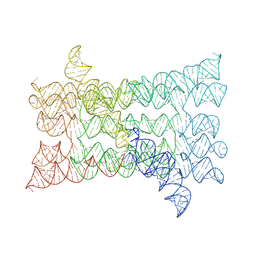
7PTK
 
 | |
5O3Y
 
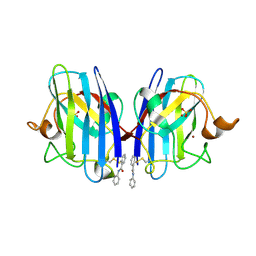 | | SOD1 bound to Ebsulfur | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Capper, M.J, Wright, G.S.A, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation.
Nat Commun, 9, 2018
|
|
8RD4
 
 | | Telomeric RAP1:DNA-PK complex | | Descriptor: | DNA (41-MER), DNA-dependent protein kinase catalytic subunit, Telomeric repeat-binding factor 2-interacting protein 1, ... | | Authors: | Eickhoff, P, Fisher, C.E.L, Inian, O, Guettler, S, Douglas, M.E. | | Deposit date: | 2023-12-07 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Chromosome end protection by RAP1-mediated inhibition of DNA-PK.
Nature, 2025
|
|
6RKO
 
 | | Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Safarian, S, Hahn, A, Kuehlbrandt, W, Michel, H. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Active site rearrangement and structural divergence in prokaryotic respiratory oxidases.
Science, 366, 2019
|
|
8SBI
 
 | |
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1R2X
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
6RS6
 
 | | X-ray crystal structure of LsAA9B | | Descriptor: | AA9, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
6RM2
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
