9FIF
 
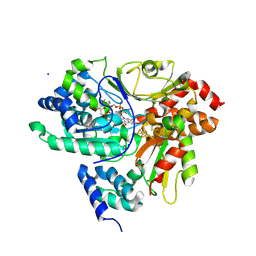 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
6QYL
 
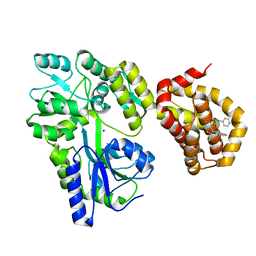 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8YK7
 
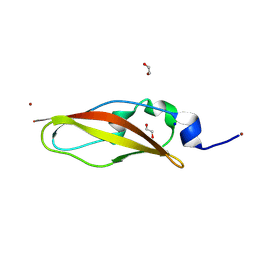 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8VMX
 
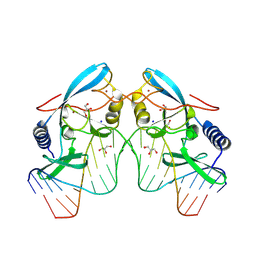 | |
8YSV
 
 | | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase, ETHANOL, ... | | Authors: | Wang, W.Y, Li, Y.J, Liu, Z.Y, Han, X.D. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis
To Be Published
|
|
9B7C
 
 | |
8VN6
 
 | |
9FCG
 
 | | Medicago truncatula 5'-ProFAR isomerase (HISN3) D57N mutant in complex with PrFAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic, ... | | Authors: | Witek, W, Imiolczyk, B, Ruszkowski, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural, kinetic, and evolutionary peculiarities of HISN3, a plant 5'-ProFAR isomerase.
Plant Physiol Biochem., 215, 2024
|
|
8VNG
 
 | |
8BYI
 
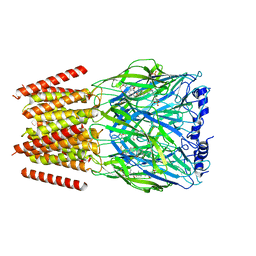 | | Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in complex with CHAPS(Alpo4_CHAPS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Acetylcholine receptor, ... | | Authors: | De Gieter, S, Efremov, R.G, Ulens, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sterol derivative binding to the orthosteric site causes conformational changes in an invertebrate Cys-loop receptor.
Elife, 12, 2023
|
|
8XF9
 
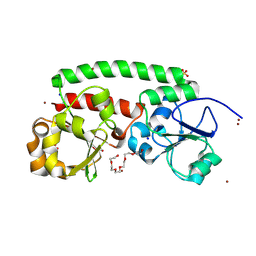 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Conserved binding mechanism for ligand promiscuity in the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes
Structure
|
|
9FE5
 
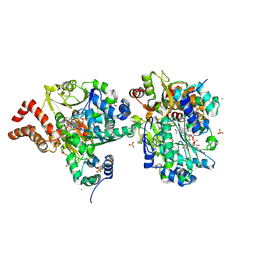 | | Crystal Structure of NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIL
 
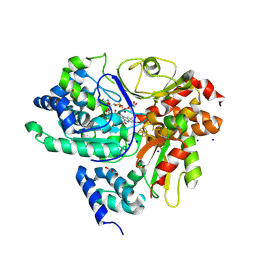 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9BJ1
 
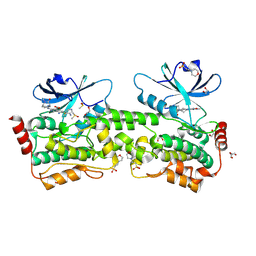 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
8VNR
 
 | |
9FQH
 
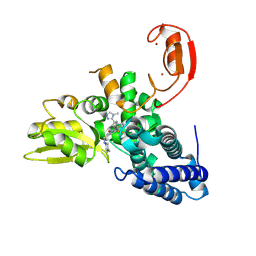 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | Descriptor: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8VUI
 
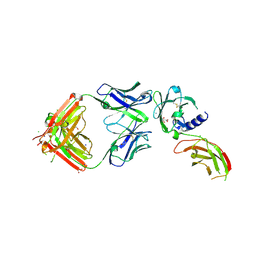 | | Structure of FabS1CE-EPR-1, an elbow-locked Fab, in complex with the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | Descriptor: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2024-03-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 22, 2024
|
|
8VMR
 
 | |
8W2C
 
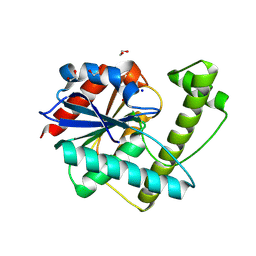 | |
8VN2
 
 | |
9C4I
 
 | | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Nascimento, K.S, Pinto-Junior, V.R, Lima, F.E.O, Osterne, V.J.S, Oliveira, M.V, Ferreira, V.M.S, Cavada, B.S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe
To Be Published
|
|
8XPE
 
 | | Crystal structure of Tris-bound TsaBgl (DATA III) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8VIG
 
 | | EgtB-IV from Geminocystis sp. isolate SKYG4, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with hercynine | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N,N-trimethyl-histidine, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
