6GOA
 
 | |
5Z7B
 
 | |
5ZUH
 
 | |
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
5BYY
 
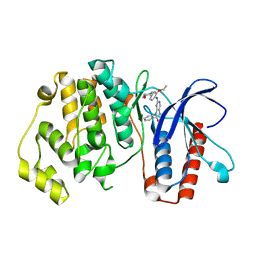 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
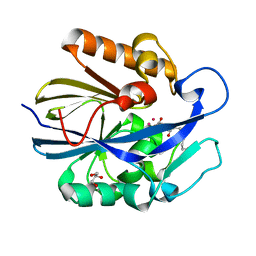
7L5F
 
 | |
1XFT
 
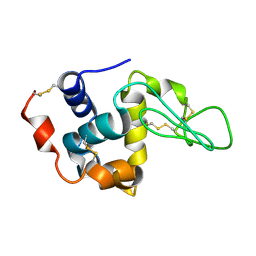 | |
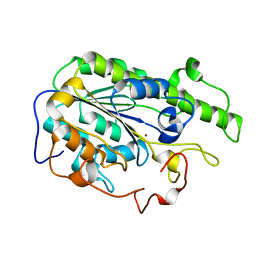
6A83
 
 | |
5WT4
 
 | | L-Cysteine-PLP intermediate of NifS from Helicobacter pylori | | Descriptor: | Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Fujishiro, T, Nakamura, R, Takahashi, T. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
6A9U
 
 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6AEQ
 
 | |
6A9V
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6AH6
 
 | | M500V mutant of Coronin coiled coil domain | | Descriptor: | Coronin-like protein | | Authors: | Ansari, A, Karade, S.S, Pratap, J.V. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6ADO
 
 | | LdCoroCC mutant-I486A | | Descriptor: | Coronin-like protein | | Authors: | Karade, S.S, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-08-01 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6A82
 
 | |
2YZ3
 
 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
1Y4O
 
 | | Solution structure of a mouse cytoplasmic Roadblock/LC7 dynein light chain | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Song, J, Tyler, R.C, Lee, M.S, Tyler, E.M, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of isoform 1 of Roadblock/LC7, a light chain in the dynein complex.
J.Mol.Biol., 354, 2005
|
|
6PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420314 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(S(-)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-31 | | Release date: | 1999-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
3I3Z
 
 | | Human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Cuprov-Netochin, R.N, Samigina, V.R, Kuranova, I.P. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5DXV
 
 | | Crystal structure of Rethreaded DHFR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Faham, S. | | Deposit date: | 2015-09-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein rethreading: A novel approach to protein design.
Sci Rep, 6, 2016
|
|
7BR9
 
 | | Crystal structure of mus musculus IRG1 | | Descriptor: | Cis-aconitate decarboxylase | | Authors: | Park, H.H, Chun, H.L. | | Deposit date: | 2020-03-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of mouse IRG1 suggests that cis-aconitate decarboxylase has an open and closed conformation.
Plos One, 15, 2020
|
|
1WD0
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*AP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7XAX
 
 | | Crystal structure of SARS coronavirus main protease in complex with Baicalei | | Descriptor: | 3C-like proteinase nsp5, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-03-19 | | Release date: | 2023-03-22 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of SARS-CoV 3C-like protease with baicalein.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
5NPU
 
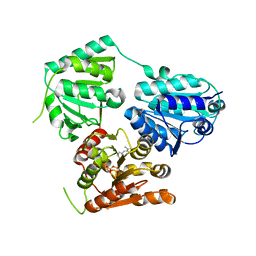 | | Inferred ancestral pyruvate decarboxylase | | Descriptor: | ANC27, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of an inferred ancestral bacterial pyruvate decarboxylase.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6FCB
 
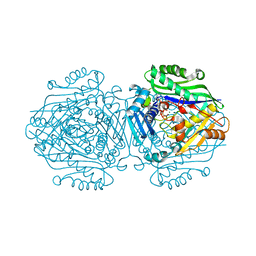 | | Human Methionine Adenosyltransferase II mutant (P115G) | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Panmanee, J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory beta subunit and quinolone-based compounds.
Febs J., 286, 2019
|
|
