2AB4
 
 | |
4XLS
 
 | |
7Q4U
 
 | |
8R6U
 
 | | Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), RNA primer, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8GY6
 
 | |
7BOE
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6MUA
 
 | |
8QU5
 
 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8HCG
 
 | | Crystal structure of mTREX1-dAMP complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCD
 
 | | Crystal structure of mTREX1-DNA product complex (dNMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCE
 
 | | Crystal structure of mTREX1-CMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCF
 
 | | Crystal structure of mTREX1-UMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Three-prime repair exonuclease 1, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCH
 
 | | Crystal structure of mTREX1-Uridine complex | | Descriptor: | MAGNESIUM ION, Three-prime repair exonuclease 1, URIDINE | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
3OIJ
 
 | |
6H02
 
 | |
6X46
 
 | | NMR solution structure of Asterix/Gtsf1 from mouse (CHHC zinc finger domains) | | Descriptor: | Gametocyte-specific factor 1, ZINC ION | | Authors: | Ipsaro, J.J, O'Brien, P.A, Bhattacharya, S, Palmer III, A.G, Joshua-Tor, L. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.
Cell Rep, 34, 2021
|
|
8PEL
 
 | |
7MLB
 
 | |
7MLI
 
 | |
7MLJ
 
 | |
2CQH
 
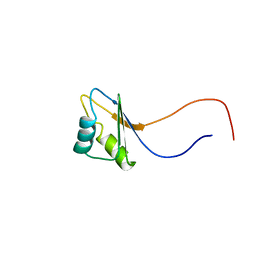 | | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2 | | Descriptor: | IGF-II mRNA-binding protein 2 isoform a | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2
To be Published
|
|
3CCU
 
 | |
3CCL
 
 | |
3CCV
 
 | |
3CCE
 
 | |
